Contribution of HLA class I (A, B, C) and HLA class II (DRB1, DQA1, DQB1) alleles and haplotypes in exploring ethnic origin of central Tunisians
- PMID: 38424564
- PMCID: PMC10903062
- DOI: 10.1186/s12920-024-01821-x
Contribution of HLA class I (A, B, C) and HLA class II (DRB1, DQA1, DQB1) alleles and haplotypes in exploring ethnic origin of central Tunisians
Abstract
Background: Estimation of HLA (Human leukocyte Antigen) alleles' frequencies in populations is essential to explore their ethnic origin. Anthropologic studies of central Tunisian population were rarely reported. Then, in this work, we aimed to explore the origin of central Tunisian population using HLA alleles and haplotypes frequencies.
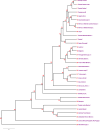
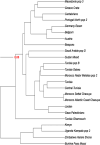
Methods: HLA class I (A, B, C) and HLA class II (DRB1, DQA1, DQB1) loci genotyping of 272 healthy unrelated organ donors was performed by Polymerase Chain Reaction-Sequence Specific Oligonucleotide (PCR-SSO). We compared central Tunisians with other populations (Arabs, Berbers, Mediterraneans, Europeans, Africans, etc.) using alleles and haplotypes frequencies, genetic distances, Neighbour-Joining dendrogram and correspondence analysis.
Results: Among the 19 HLA A alleles, the 26 HLA B alleles, the 13 HLA C alleles, the 15 HLA DRB1 alleles, the 6 HLA DQA1 alleles and the 5 HLA DQB1 alleles identified in the studied population, HLA A*02 (22.8%), HLA B*50 (13.1%), HLA C*06 (21.8%), HLA DRB1*07 (17.8%), HLA DQA1*01 (32.1%) and HLA DQB1*03 (31.6%) were the most frequent alleles. The extended haplotypes HLA A*02-B*50-C*06-DRB1*07-DQA1*02-DQB1*02 (1.97%) was the most frequent HLA six-loci haplotype.
Conclusion: Central Tunisians were very close to other Tunisian populations, to Iberians and North Africans. They were rather distant from sub-Saharan populations and eastern Mediterraneans especially Arabs although the strong cultural and religious impact of Arabs in this population.
Keywords: Anthropology; Genetic polymorphism; Haplotypes; Human leukocyte Antigen (HLA).
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Molecular analysis of HLA allelic frequencies and haplotypes in Jordanians and comparison with other related populations.Hum Immunol. 2001 Sep;62(9):901-9. doi: 10.1016/s0198-8859(01)00289-0. Hum Immunol. 2001. PMID: 11543892
-
The investigation of the origin of Southern Tunisians using HLA genes.J Hum Genet. 2017 Mar;62(3):419-429. doi: 10.1038/jhg.2016.146. Epub 2016 Nov 24. J Hum Genet. 2017. PMID: 27881842
-
HLA class I (A, B, C) and class II (DRB1, DQA1, DQB1, DPB1) alleles and haplotypes in the Han from southern China.Tissue Antigens. 2007 Dec;70(6):455-63. doi: 10.1111/j.1399-0039.2007.00932.x. Epub 2007 Sep 27. Tissue Antigens. 2007. PMID: 17900288
-
HLA Class II Allele and Haplotype Diversity in Libyans and Their Genetic Relationships with Other Populations.Immunol Invest. 2019 Nov;48(8):875-892. doi: 10.1080/08820139.2019.1614950. Epub 2019 Jun 4. Immunol Invest. 2019. PMID: 31161824
-
Genetic Diversity and Ethnic Tapestry of Kazakhstan as Inferred from HLA Polymorphism and Population Dynamics: A Comprehensive Review.Genes (Basel). 2025 Mar 15;16(3):342. doi: 10.3390/genes16030342. Genes (Basel). 2025. PMID: 40149493 Free PMC article. Review.
Cited by
-
High resolution class I HLA-A, -B, and -C diversity in Eastern and Southern African populations.Sci Rep. 2025 Jul 2;15(1):23667. doi: 10.1038/s41598-025-06704-4. Sci Rep. 2025. PMID: 40603473 Free PMC article.
-
High Resolution Class I HLA -A, -B, and -C Diversity in Eastern and Southern African Populations.bioRxiv [Preprint]. 2024 Sep 8:2024.09.04.611164. doi: 10.1101/2024.09.04.611164. bioRxiv. 2024. Update in: Sci Rep. 2025 Jul 2;15(1):23667. doi: 10.1038/s41598-025-06704-4. PMID: 39282263 Free PMC article. Updated. Preprint.
References
-
- Shiina T, Hosomichi K, Inoko H, Kulski JK. The HLA genomic loci map: expression, interaction, diversity and disease. J Hum Genet. 2009;54(1):15–39. - PubMed
-
- Riley E, Olerup O. HLA polymorphisms and evolution. Immunol Today. 1992;13(9):333–335. - PubMed
-
- Abdennaji Guenounou B, Loueslati BY, Buhler S, Hmida S, Ennafaa H, Khodjet-Elkhil H, et al. HLA class II genetic diversity in southern Tunisia and the Mediterranean area. Int J Immunogenet. 2006;33(2):93–103. - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Research Materials

