New insights into bioaugmented removal of sulfamethoxazole in sediment microcosms: degradation efficiency, ecological risk and microbial mechanisms
- PMID: 38424602
- PMCID: PMC10903153
- DOI: 10.1186/s40168-023-01741-5
New insights into bioaugmented removal of sulfamethoxazole in sediment microcosms: degradation efficiency, ecological risk and microbial mechanisms
Abstract
Background: Bioaugmentation has the potential to enhance the ability of ecological technology to treat sulfonamide-containing wastewater, but the low viability of the exogenous degraders limits their practical application. Understanding the mechanism is important to enhance and optimize performance of the bioaugmentation, which requires a multifaceted analysis of the microbial communities. Here, DNA-stable isotope probing (DNA-SIP) and metagenomic analysis were conducted to decipher the bioaugmentation mechanisms in stabilization pond sediment microcosms inoculated with sulfamethoxazole (SMX)-degrading bacteria (Pseudomonas sp. M2 or Paenarthrobacter sp. R1).
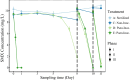
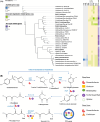
Results: The bioaugmentation with both strains M2 and R1, especially strain R1, significantly improved the biodegradation rate of SMX, and its biodegradation capacity was sustainable within a certain cycle (subjected to three repeated SMX additions). The removal strategy using exogenous degrading bacteria also significantly abated the accumulation and transmission risk of antibiotic resistance genes (ARGs). Strain M2 inoculation significantly lowered bacterial diversity and altered the sediment bacterial community, while strain R1 inoculation had a slight effect on the bacterial community and was closely associated with indigenous microorganisms. Paenarthrobacter was identified as the primary SMX-assimilating bacteria in both bioaugmentation systems based on DNA-SIP analysis. Combining genomic information with pure culture evidence, strain R1 enhanced SMX removal by directly participating in SMX degradation, while strain M2 did it by both participating in SMX degradation and stimulating SMX-degrading activity of indigenous microorganisms (Paenarthrobacter) in the community.
Conclusions: Our findings demonstrate that bioaugmentation using SMX-degrading bacteria was a feasible strategy for SMX clean-up in terms of the degradation efficiency of SMX, the risk of ARG transmission, as well as the impact on the bacterial community, and the advantage of bioaugmentation with Paenarthrobacter sp. R1 was also highlighted. Video Abstract.
Keywords: Antibiotic resistance genes (ARGs); Bioaugmentation; DNA-stable isotope probing; Metagenomics; Sulfonamide.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
The presence of in situ sulphamethoxazole degraders and their interactions with other microbes in activated sludge as revealed by DNA stable isotope probing and molecular ecological network analysis.Environ Int. 2019 Mar;124:121-129. doi: 10.1016/j.envint.2018.12.039. Epub 2019 Jan 11. Environ Int. 2019. PMID: 30641255
-
Anaerobic sulfamethoxazole-degrading bacterial consortia in antibiotic-contaminated wetland sediments identified by DNA-stable isotope probing and metagenomics analysis.Environ Microbiol. 2022 Aug;24(8):3751-3763. doi: 10.1111/1462-2920.16091. Epub 2022 Jun 10. Environ Microbiol. 2022. PMID: 35688651
-
The sulfonamide-resistance dihydropteroate synthase gene is crucial for efficient biodegradation of sulfamethoxazole by Paenarthrobacter species.Appl Microbiol Biotechnol. 2023 Sep;107(18):5813-5827. doi: 10.1007/s00253-023-12679-x. Epub 2023 Jul 13. Appl Microbiol Biotechnol. 2023. PMID: 37439835
-
Microbial degradation of sulfamethoxazole in the environment.Appl Microbiol Biotechnol. 2018 Apr;102(8):3573-3582. doi: 10.1007/s00253-018-8845-4. Epub 2018 Mar 7. Appl Microbiol Biotechnol. 2018. PMID: 29516143 Review.
-
Biodegradation of phenolic pollutants and bioaugmentation strategies: A review of current knowledge and future perspectives.J Hazard Mater. 2024 May 5;469:133906. doi: 10.1016/j.jhazmat.2024.133906. Epub 2024 Feb 28. J Hazard Mater. 2024. PMID: 38430590 Review.
Cited by
-
Cross-feeding and co-degradation within a bacterial consortium dominated by challenging-to-culture Leucobacter sp. HA-1 enhances sulfonamide degradation.Appl Environ Microbiol. 2025 Jul 23;91(7):e0059025. doi: 10.1128/aem.00590-25. Epub 2025 Jun 24. Appl Environ Microbiol. 2025. PMID: 40552827 Free PMC article.
References
-
- Qiao M, Ying G, Singer AC, Zhu Y. Review of antibiotic resistance in China and its environment. Environ Int. 2018;110:160–172. - PubMed
-
- Deng Y, Li B, Zhang T. Bacteria that make a meal of sulfonamide antibiotics: blind spots and emerging opportunities. Environ Sci Technol. 2018;52:3854–3868. - PubMed
-
- Zhou L, Wu QL, Zhang B, Zhao Y, Zhao B. Occurrence, spatiotemporal distribution, mass balance and ecological risks of antibiotics in subtropical shallow Lake Taihu, China. Environ Sci Process Impacts. 2016;18:500–513. - PubMed
-
- Guo X, Feng C, Gu E, Tian C, Shen Z. Spatial distribution, source apportionment and risk assessment of antibiotics in the surface water and sediments of the Yangtze Estuary. Sci Total Environ. 2019;671:548–557. - PubMed
-
- Spielmeyer A, Höper H, Hamscher G. Long-term monitoring of sulfonamide leaching from manure amended soil into groundwater. Chemosphere. 2017;177:232–238. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

