Construction of a versatile in vitro cultivation screening platform using human oral microbiota
- PMID: 38425145
- PMCID: PMC10904971
- DOI: 10.1111/1758-2229.13243
Construction of a versatile in vitro cultivation screening platform using human oral microbiota
Abstract
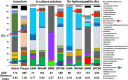
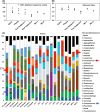
We developed a simulation model of human oral microbiota using Bio Palette oral medium (BPOM) containing 0.02% glucose and lower bacterial nitrogen sources, derived from saliva and dental plaque. By decreasing the concentration of Gifu anaerobic medium (GAM) from 30 to 10 g L-1 , we observed increased ratios of target pathogenic genera, Porphyromonas and Fusobacterium from 0.5% and 1.7% to 1.2% and 3.5%, respectively, in the biofilm on hydroxyapatite (HA) discs. BPOM exhibited the higher ratios of Porphyromonas and Fusobacterium, and amplicon sequence variant number on HA, compared with GAM, modified GAM and basal medium mucin. Mixing glycerol stocks of BPOM culture solutions from four human subjects resulted in comparable ratios of these bacteria to the original saliva. In this simulation model, sitafloxacin showed higher inhibitory effects on P. gingivalis than minocycline hydrochloride at a low dosage of 0.1 μg mL-1 . Probiotics such as Streptococcus salivarius and Limosilactobacillus fermentum also showed significant decreases in Porphyromonas and Fusobacterium ratios on HA, respectively. Overall, the study suggests that BPOM with low carbon and nutrients could be a versatile platform for assessing the efficacy of antibiotics and live biotherapeutics in treating oral diseases caused by Porphyromonas and Fusobacterium.
© 2024 The Authors. Environmental Microbiology Reports published by Applied Microbiology International and John Wiley & Sons Ltd.
Conflict of interest statement
Kengo Sasaki, Yasunobu Takeshima, Ayami Fujino and Ryo Okumura are employees of Bio Palette Co., Ltd. All other authors declare no competing interests.
Figures


Similar articles
-
Fusobacterium nucleatum Metabolically Integrates Commensals and Pathogens in Oral Biofilms.mSystems. 2022 Aug 30;7(4):e0017022. doi: 10.1128/msystems.00170-22. Epub 2022 Jul 19. mSystems. 2022. PMID: 35852319 Free PMC article.
-
Label-free quantitative proteomic analysis of the oral bacteria Fusobacterium nucleatum and Porphyromonas gingivalis to identify protein features relevant in biofilm formation.Anaerobe. 2021 Dec;72:102449. doi: 10.1016/j.anaerobe.2021.102449. Epub 2021 Sep 17. Anaerobe. 2021. PMID: 34543761
-
Antimicrobial photodynamic therapy alone or in combination with antibiotic local administration against biofilms of Fusobacterium nucleatum and Porphyromonas gingivalis.J Photochem Photobiol B. 2018 Nov;188:135-145. doi: 10.1016/j.jphotobiol.2018.09.010. Epub 2018 Sep 12. J Photochem Photobiol B. 2018. PMID: 30267963
-
Can metagenomics unravel the impact of oral bacteriome in human diseases?Biotechnol Genet Eng Rev. 2023 Apr;39(1):85-117. doi: 10.1080/02648725.2022.2102877. Epub 2022 Jul 21. Biotechnol Genet Eng Rev. 2023. PMID: 35861776 Review.
-
Role of Porphyromonas gingivalis in oral and orodigestive squamous cell carcinoma.Periodontol 2000. 2022 Jun;89(1):154-165. doi: 10.1111/prd.12425. Epub 2022 Mar 4. Periodontol 2000. 2022. PMID: 35244980 Free PMC article. Review.
Cited by
-
Effect of Mouth Rinsing and Antiseptic Solutions on Periodontitis Bacteria in an In Vitro Oral Human Biofilm Model.Dent J (Basel). 2025 Jul 16;13(7):324. doi: 10.3390/dj13070324. Dent J (Basel). 2025. PMID: 40710169 Free PMC article.
-
The rheumatoid arthritis gut microbial biobank reveals core microbial species that associate and effect on host inflammation and autoimmune responses.Imeta. 2024 Oct 3;3(5):e242. doi: 10.1002/imt2.242. eCollection 2024 Oct. Imeta. 2024. PMID: 39429876 Free PMC article.
References
-
- Amano, A. , Kuboniwa, M. , Nakagawa, I. , Akiyama, S. , Morisaki, I. & Hamada, S. (2000) Prevalence of specific genotypes of Porphyromonas gingivalis fimA and periodontal health status. Journal of Dental Research, 79, 1664–1668. - PubMed
-
- Brown, J.L. , Johnston, W. , Delaney, C. , Short, B. , Butcher, M.C. , Young, T. et al. (2019) Polymicrobial oral biofilm models: simplifying the complex. Journal of Medical Microbiology, 68, 1573–1584. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

