MtDNA deletions and aging
- PMID: 38425363
- PMCID: PMC10902006
- DOI: 10.3389/fragi.2024.1359638
MtDNA deletions and aging
Abstract
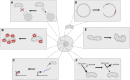
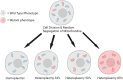
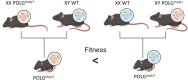
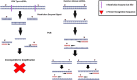
Aging is the major risk factor in most of the leading causes of mortality worldwide, yet its fundamental causes mostly remain unclear. One of the clear hallmarks of aging is mitochondrial dysfunction. Mitochondria are best known for their roles in cellular energy generation, but they are also critical biosynthetic and signaling organelles. They also undergo multiple changes with organismal age, including increased genetic errors in their independent, circular genome. A key group of studies looking at mice with increased mtDNA mutations showed that premature aging phenotypes correlated with increased deletions but not point mutations. This generated an interest in mitochondrial deletions as a potential fundamental cause of aging. However, subsequent studies in different models have yielded diverse results. This review summarizes the research on mitochondrial deletions in various organisms to understand their possible roles in causing aging while identifying the key complications in quantifying deletions across all models.
Keywords: aging; mitochondria; mitochondrial DNA; mitochondrial DNA deletions; mitochondrial dysfunction.
Copyright © 2024 Sprason, Tucker and Clancy.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The authors declared that they were editorial board members of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures

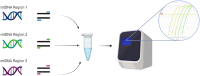
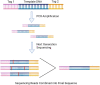
References
-
- Allio R., Donega S., Galtier N., Nabholz B. (2017). Large variation in the ratio of mitochondrial to nuclear mutation rate across animals: implications for genetic diversity and the use of mitochondrial DNA as a molecular marker. Mol. Biol. Evol. 34 (11), 2762–2772. 10.1093/molbev/msx197 - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources

