Early results of the integrative epigenomic-transcriptomic landscape of colorectal adenoma and cancer
- PMID: 38425399
- PMCID: PMC10900154
- DOI: 10.4251/wjgo.v16.i2.414
Early results of the integrative epigenomic-transcriptomic landscape of colorectal adenoma and cancer
Abstract
Background: Aberrant methylation is common during the initiation and progression of colorectal cancer (CRC), and detecting these changes that occur during early adenoma (ADE) formation and CRC progression has clinical value.
Aim: To identify potential DNA methylation markers specific to ADE and CRC.
Methods: Here, we performed SeqCap targeted bisulfite sequencing and RNA-seq analysis of colorectal ADE and CRC samples to profile the epigenomic-transcriptomic landscape.
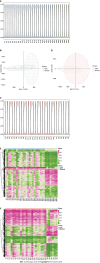
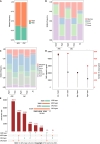
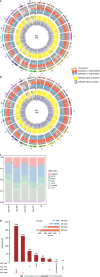
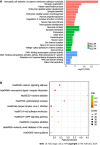
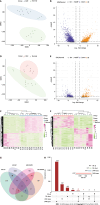
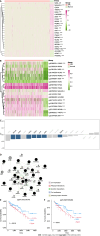
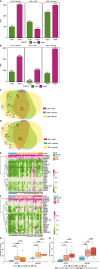
Results: Comparing 22 CRC and 25 ADE samples, global methylation was higher in the former, but both showed similar methylation patterns regarding differentially methylated gene positions, chromatin signatures, and repeated elements. High-grade CRC tended to exhibit elevated methylation levels in gene promoter regions compared to those in low-grade CRC. Combined with RNA-seq gene expression data, we identified 14 methylation-regulated differentially expressed genes, of which only AGTR1 and NECAB1 methylation had prognostic significance.
Conclusion: Our results suggest that genome-wide alterations in DNA methylation occur during the early stages of CRC and demonstrate the methylation signatures associated with colorectal ADEs and CRC, suggesting prognostic biomarkers for CRC.
Keywords: Colorectal cancer; Epigenomic alteration; Methylation-regulated differentially expressed genes; Transcriptome.
©The Author(s) 2024. Published by Baishideng Publishing Group Inc. All rights reserved.
Conflict of interest statement
Conflict-of-interest statement: All the authors report no relevant conflicts of interest for this article.
Figures

References
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209–249. - PubMed
-
- Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. - PubMed
LinkOut - more resources
Full Text Sources

