Epilepsy and sudden unexpected death in epilepsy in a mouse model of human SCN1B-linked developmental and epileptic encephalopathy
- PMID: 38425576
- PMCID: PMC10903178
- DOI: 10.1093/braincomms/fcad283
Epilepsy and sudden unexpected death in epilepsy in a mouse model of human SCN1B-linked developmental and epileptic encephalopathy
Erratum in
-
Correction to: Epilepsy and sudden unexpected death in epilepsy in a mouse model of human SCN1B-linked developmental and epileptic encephalopathy.Brain Commun. 2024 Jan 24;6(1):fcae009. doi: 10.1093/braincomms/fcae009. eCollection 2024. Brain Commun. 2024. PMID: 38274569 Free PMC article.
Abstract
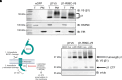
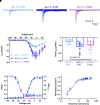
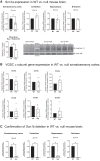
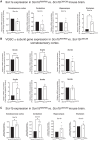
Voltage-gated sodium channel β1 subunits are essential proteins that regulate excitability. They modulate sodium and potassium currents, function as cell adhesion molecules and regulate gene transcription following regulated intramembrane proteolysis. Biallelic pathogenic variants in SCN1B, encoding β1, are linked to developmental and epileptic encephalopathy 52, with clinical features overlapping Dravet syndrome. A recessive variant, SCN1B-c.265C>T, predicting SCN1B-p.R89C, was homozygous in two children of a non-consanguineous family. One child was diagnosed with Dravet syndrome, while the other had a milder phenotype. We identified an unrelated biallelic SCN1B-c.265C>T patient with a clinically more severe phenotype than Dravet syndrome. We used CRISPR/Cas9 to knock-in SCN1B-p.R89C to the mouse Scn1b locus (Scn1bR89/C89). We then rederived the line on the C57BL/6J background to allow comparisons between Scn1bR89/R89 and Scn1bC89/C89 littermates with Scn1b+/+ and Scn1b-/- mice, which are congenic on C57BL/6J, to determine whether the SCN1B-c.265C>T variant results in loss-of-function. Scn1bC89/C89 mice have normal body weights and ∼20% premature mortality, compared with severely reduced body weight and 100% mortality in Scn1b-/- mice. β1-p.R89C polypeptides are expressed in brain at comparable levels to wild type. In heterologous cells, β1-p.R89C localizes to the plasma membrane and undergoes regulated intramembrane proteolysis similar to wild type. Heterologous expression of β1-p.R89C results in sodium channel α subunit subtype specific effects on sodium current. mRNA abundance of Scn2a, Scn3a, Scn5a and Scn1b was increased in Scn1bC89/C89 somatosensory cortex, with no changes in Scn1a. In contrast, Scn1b-/- mouse somatosensory cortex is haploinsufficient for Scn1a, suggesting an additive mechanism for the severity of the null model via disrupted regulation of another Dravet syndrome gene. Scn1bC89/C89 mice are more susceptible to hyperthermia-induced seizures at post-natal Day 15 compared with Scn1bR89/R89 littermates. EEG recordings detected epileptic discharges in young adult Scn1bC89/C89 mice that coincided with convulsive seizures and myoclonic jerks. We compared seizure frequency and duration in a subset of adult Scn1bC89/C89 mice that had been exposed to hyperthermia at post-natal Day 15 versus a subset that were not hyperthermia exposed. No differences in spontaneous seizures were detected between groups. For both groups, the spontaneous seizure pattern was diurnal, occurring with higher frequency during the dark cycle. This work suggests that the SCN1B-c.265C>T variant does not result in complete loss-of-function. Scn1bC89/C89 mice more accurately model SCN1B-linked variants with incomplete loss-of-function compared with Scn1b-/- mice, which model complete loss-of-function, and thus add to our understanding of disease mechanisms as well as our ability to develop new therapeutic strategies.
Keywords: SUDEP; developmental and epileptic encephalopathy; seizure; sodium channel; β subunit.
© The Author(s) 2023. Published by Oxford University Press on behalf of the Guarantors of Brain.
Conflict of interest statement
The authors report no competing interests.
Figures




References
-
- Dravet C, Bureau M, Oguni H, Fukuyama Y, Cokar O. Severe myoclonic epilepsy in infancy (Dravet syndrome). Adv Neurol. 2005;95:71–102. - PubMed
-
- Guerrini R, Aicardi J. Epileptic encephalopathies with myoclonic seizures in infants and children (severe myoclonic epilepsy and myoclonic-astatic epilepsy). J Clin Neurophysiol. 2003;20(6):449–461. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
