Deregulation of circRNA hsa_circ_0009109 promotes tumor growth and initiates autophagy by sponging miR-544a-3p in gastric cancer
- PMID: 38425655
- PMCID: PMC10902679
- DOI: 10.1093/gastro/goae008
Deregulation of circRNA hsa_circ_0009109 promotes tumor growth and initiates autophagy by sponging miR-544a-3p in gastric cancer
Abstract
Background: Autophagy death of cancer cells is detrimental to apoptosis induced by therapeutic drugs, which promotes tumor progression to a certain extent. Increasing reports have demonstrated the regulatory role of circular RNAs (circRNAs) in autophagy. Here, we aimed to determine the role of hsa_circ_0009109 in autophagy in gastric cancer (GC).
Methods: The effects of hsa_circ_0009109 on autophagy were examined using quantitative real-time polymerase chain reaction (qPCR), transmission electron microscopy, Western blot, and immunofluorescence. The mechanism of hsa_circ_0009109 regulating the miR-544a-3p/bcl-2 axis was analysed using fluorescence in situ hybridization, dual-luciferase reporter, and rescue experiments.
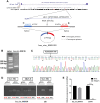
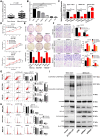
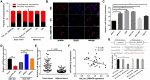
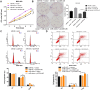
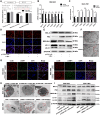
Results: Functional testing indicated that hsa_circ_0009109 was significantly down-expressed in GC tissues and cell lines. A reduction in cytoplasmic-derived hsa_circ_0009109 could promote GC progression by accelerating cell proliferation, enhancing migration and invasion, inhibiting apoptosis, and accelerating the cell cycle progression. Besides, hsa_circ_0009109 was found to exert the effect of an autophagy inhibitor such as 3-Methyladenine (3-MA), which was manifested by the weakening of the immunofluorescence of LC3B and the reduction in autophagy-related proteins after overexpression of hsa_circ_0009109, while increased autophagosomes were observed after interference with hsa_circ_0009109. Subsequently, the crosstalk between hsa_circ_0009109 and miR-544a-3p/bcl-2 was verified using dual-luciferase reporter assay. The autophagy status was altered under the regulation of the hsa_circ_0009109-targeted miR-544a-3p/bcl-2 axis.
Conclusions: The hsa_circ_0009109 mediated a novel autophagy regulatory network through targeting the miR-544a-3p/bcl-2 axis, which may shed new light on the exploration of therapeutic targets for the clinical treatment of GC.
Keywords: autophagy; circRNA; gastric cancer; hsa_circ_0009109; miR-544a-3p.
© The Author(s) 2024. Published by Oxford University Press and Sixth Affiliated Hospital of Sun Yat-sen University.
Conflict of interest statement
The authors declare that there is no conflict of interests in this study.
Figures

Similar articles
-
Hsa_circ_RNA_0011780 Represses the Proliferation and Metastasis of Non-Small Cell Lung Cancer by Decreasing FBXW7 via Targeting miR-544a.Onco Targets Ther. 2020 Jan 23;13:745-755. doi: 10.2147/OTT.S236162. eCollection 2020. Onco Targets Ther. 2020. Retraction in: Onco Targets Ther. 2022 Nov 16;15:1385-1386. doi: 10.2147/OTT.S397666. PMID: 32158226 Free PMC article. Retracted.
-
Hsa_circ_0003159 inhibits gastric cancer progression by regulating miR-223-3p/NDRG1 axis.Cancer Cell Int. 2020 Feb 19;20:57. doi: 10.1186/s12935-020-1119-0. eCollection 2020. Cancer Cell Int. 2020. PMID: 32099530 Free PMC article.
-
circRNA_0006470 promotes the proliferation and migration of gastric cancer cells by functioning as a sponge of miR-27b-3p.Neoplasma. 2021 Nov;68(6):1245-1256. doi: 10.4149/neo_2021_210222N235. Epub 2021 Oct 10. Neoplasma. 2021. PMID: 34641696
-
hsa_circ_0023409 Accelerates Gastric Cancer Cell Growth and Metastasis Through Regulating the IRS4/PI3K/AKT Pathway.Cell Transplant. 2021 Jan-Dec;30:963689720975390. doi: 10.1177/0963689720975390. Cell Transplant. 2021. Retraction in: Cell Transplant. 2024 Jan-Dec;33:9636897241298459. doi: 10.1177/09636897241298459. PMID: 33439739 Free PMC article. Retracted.
-
Hsa_circ_0088036 promotes nonsmall cell lung cancer progression by regulating miR-1343-3p/Bcl-3 axis through TGFβ/Smad3/EMT signaling.Mol Carcinog. 2023 Jul;62(7):1073-1085. doi: 10.1002/mc.23547. Epub 2023 May 3. Mol Carcinog. 2023. PMID: 37132942
Cited by
-
Research progress on N6-methyladenosine and non-coding RNA in multiple myeloma.Discov Oncol. 2025 Apr 25;16(1):615. doi: 10.1007/s12672-025-02386-6. Discov Oncol. 2025. PMID: 40281359 Free PMC article. Review.
References
-
- Sung H, Ferlay J, Siegel RL. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71:209–49. - PubMed
-
- Chen W, Zheng R, Baade PD. et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115–32. - PubMed
-
- Beermann J, Piccoli MT, Viereck J. et al. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev 2016;96:1297–325. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

