A multi-scale numerical approach to study monoclonal antibodies in solution
- PMID: 38425712
- PMCID: PMC10902793
- DOI: 10.1063/5.0186642
A multi-scale numerical approach to study monoclonal antibodies in solution
Abstract
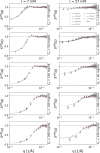
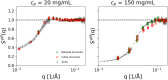
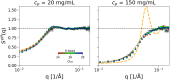
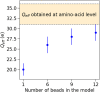
Developing efficient and robust computational models is essential to improve our understanding of protein solution behavior. This becomes particularly important to tackle the high-concentration regime. In this context, the main challenge is to put forward coarse-grained descriptions able to reduce the level of detail, while retaining key features and relevant information. In this work, we develop an efficient strategy that can be used to investigate and gain insight into monoclonal antibody solutions under different conditions. We use a multi-scale numerical approach, which connects information obtained at all-atom and amino-acid levels to bead models. The latter has the advantage of reproducing the properties of interest while being computationally much faster. Indeed, these models allow us to perform many-protein simulations with a large number of molecules. We can, thus, explore conditions not easily accessible with more detailed descriptions, perform effective comparisons with experimental data up to very high protein concentrations, and efficiently investigate protein-protein interactions and their role in phase behavior and protein self-assembly. Here, a particular emphasis is given to the effects of charges at different ionic strengths.
© 2024 Author(s).
Conflict of interest statement
The authors have no conflicts to disclose.
Figures





Similar articles
-
Cross-correlation corrected friction in generalized Langevin models: Application to the continuous Asakura-Oosawa model.J Chem Phys. 2022 Jul 28;157(4):044103. doi: 10.1063/5.0093056. J Chem Phys. 2022. PMID: 35922348
-
The future of Cochrane Neonatal.Early Hum Dev. 2020 Nov;150:105191. doi: 10.1016/j.earlhumdev.2020.105191. Epub 2020 Sep 12. Early Hum Dev. 2020. PMID: 33036834
-
Coarse-Grained Antibody Models for "Weak" Protein-Protein Interactions from Low to High Concentrations.J Phys Chem B. 2016 Jul 14;120(27):6592-605. doi: 10.1021/acs.jpcb.6b04907. Epub 2016 Jul 1. J Phys Chem B. 2016. PMID: 27314827
-
Computational models for studying physical instabilities in high concentration biotherapeutic formulations.MAbs. 2022 Jan-Dec;14(1):2044744. doi: 10.1080/19420862.2022.2044744. MAbs. 2022. PMID: 35282775 Free PMC article. Review.
-
Spectroscopic methods for assessing the molecular origins of macroscopic solution properties of highly concentrated liquid protein solutions.Anal Biochem. 2018 Nov 15;561-562:70-88. doi: 10.1016/j.ab.2018.09.013. Epub 2018 Sep 20. Anal Biochem. 2018. PMID: 30243977 Review.
Cited by
-
Guest Editorial: Structure and mechanics of biofluids, biomaterials, and biologics.APL Bioeng. 2025 May 27;9(2):020401. doi: 10.1063/5.0274572. eCollection 2025 Jun. APL Bioeng. 2025. PMID: 40438387 Free PMC article. No abstract available.
-
Combining Scattering Experiments and Colloid Theory to Characterize Charge Effects in Concentrated Antibody Solutions.Mol Pharm. 2024 May 6;21(5):2250-2271. doi: 10.1021/acs.molpharmaceut.3c01023. Epub 2024 Apr 25. Mol Pharm. 2024. PMID: 38661388 Free PMC article.
-
Harnessing computational technologies to facilitate antibody-drug conjugate development.Nat Chem Biol. 2025 Aug;21(8):1138-1147. doi: 10.1038/s41589-025-01950-z. Epub 2025 Jun 27. Nat Chem Biol. 2025. PMID: 40579571 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources

