Proteolysis Targeting Chimera Degraders of the METTL3-14 m6A-RNA Methyltransferase
- PMID: 38425900
- PMCID: PMC10900215
- DOI: 10.1021/jacsau.4c00040
Proteolysis Targeting Chimera Degraders of the METTL3-14 m6A-RNA Methyltransferase
Abstract
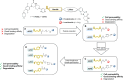
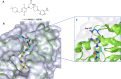
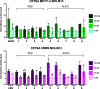
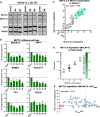
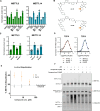
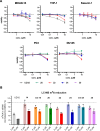
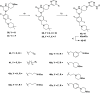
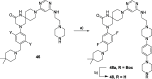
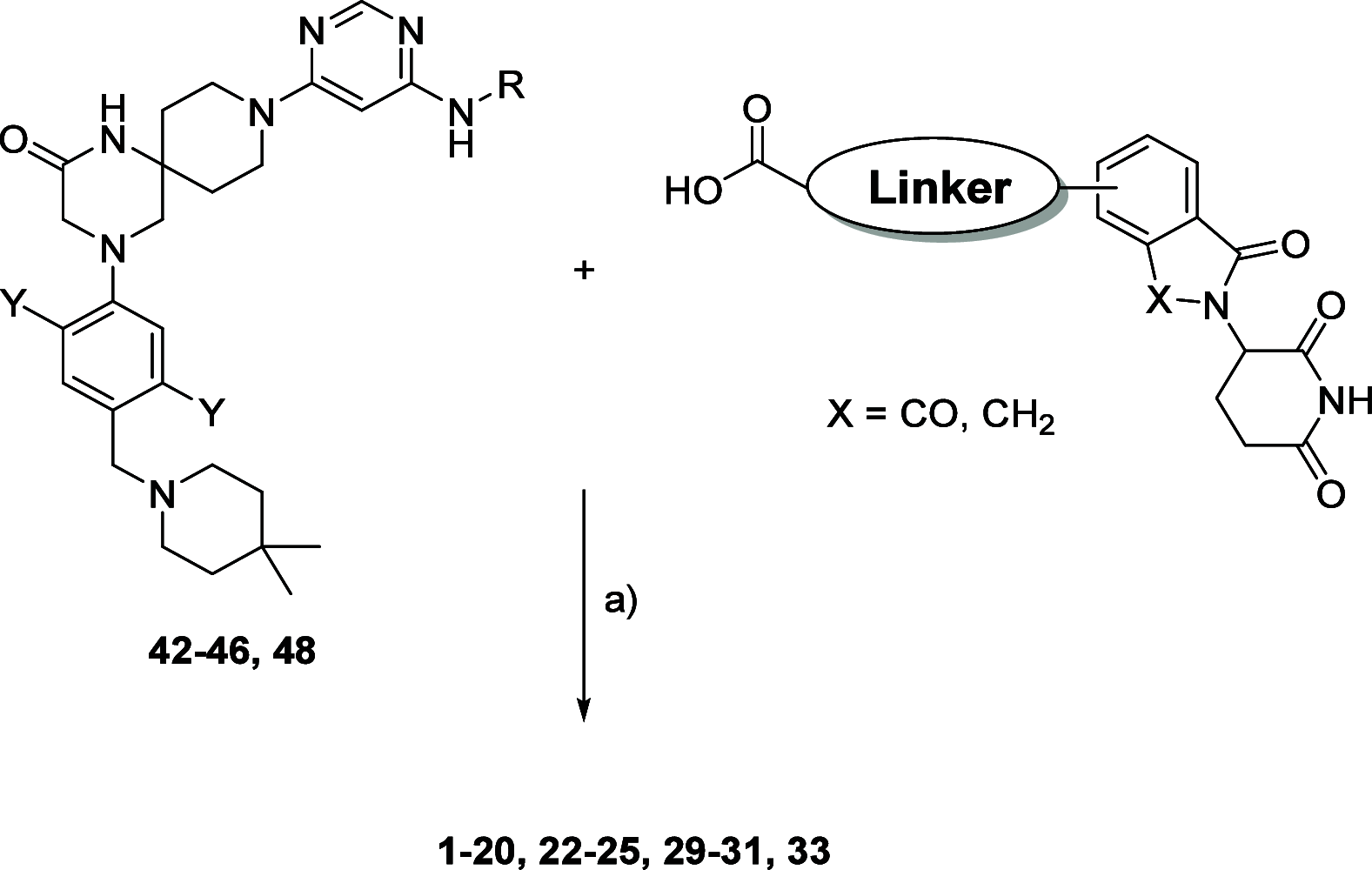
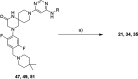
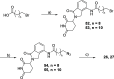
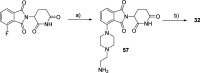
Methylation of adenine N6 (m6A) is the most frequent RNA modification. On mRNA, it is catalyzed by the METTL3-14 heterodimer complex, which plays a key role in acute myeloid leukemia (AML) and other types of blood cancers and solid tumors. Here, we disclose the first proteolysis targeting chimeras (PROTACs) for an epitranscriptomics protein. For designing the PROTACs, we made use of the crystal structure of the complex of METTL3-14 with a potent and selective small-molecule inhibitor (called UZH2). The optimization of the linker started from a desfluoro precursor of UZH2 whose synthesis is more efficient than that of UZH2. The first nine PROTAC molecules featured PEG- or alkyl-based linkers, but only the latter showed cell penetration. With this information in hand, we synthesized 26 PROTACs based on UZH2 and alkyl linkers of different lengths and rigidity. The formation of the ternary complex was validated by a FRET-based biochemical assay and an in vitro ubiquitination assay. The PROTACs 14, 20, 22, 24, and 30, featuring different linker types and lengths, showed 50% or higher degradation of METTL3 and/or METTL14 measured by Western blot in MOLM-13 cells. They also showed substantial degradation on three other AML cell lines and prostate cancer cell line PC3.
© 2024 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


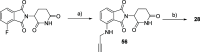

Similar articles
-
Structure-guided design of a methyltransferase-like 3 (METTL3) proteolysis targeting chimera (PROTAC) incorporating an indole-nicotinamide chemotype.RSC Med Chem. 2025 Jun 19. doi: 10.1039/d5md00359h. Online ahead of print. RSC Med Chem. 2025. PMID: 40599585 Free PMC article.
-
Targeting METTL3 protein by proteolysis-targeting chimeras: A novel therapeutic approach for acute myeloid leukemia.Genes Dis. 2024 Nov 7;12(4):101452. doi: 10.1016/j.gendis.2024.101452. eCollection 2025 Jul. Genes Dis. 2024. PMID: 40453361 Free PMC article.
-
Discovery of a PROTAC degrader for METTL3-METTL14 complex.Cell Chem Biol. 2024 Jan 18;31(1):177-183.e17. doi: 10.1016/j.chembiol.2023.12.009. Epub 2024 Jan 8. Cell Chem Biol. 2024. PMID: 38194973
-
Assays and technologies for developing proteolysis targeting chimera degraders.Future Med Chem. 2020 Jun;12(12):1155-1179. doi: 10.4155/fmc-2020-0073. Epub 2020 May 20. Future Med Chem. 2020. PMID: 32431173 Free PMC article. Review.
-
PROTACs: Current Trends in Protein Degradation by Proteolysis-Targeting Chimeras.BioDrugs. 2022 Sep;36(5):609-623. doi: 10.1007/s40259-022-00551-9. Epub 2022 Sep 13. BioDrugs. 2022. PMID: 36098871 Review.
Cited by
-
Small molecule inhibitors targeting m6A regulators.J Hematol Oncol. 2024 May 6;17(1):30. doi: 10.1186/s13045-024-01546-5. J Hematol Oncol. 2024. PMID: 38711100 Free PMC article. Review.
-
Patent landscape of small molecule inhibitors of METTL3 (2020-present).Expert Opin Ther Pat. 2024 Dec 26:1-16. doi: 10.1080/13543776.2024.2447056. Online ahead of print. Expert Opin Ther Pat. 2024. PMID: 39721070 Review.
-
Recent advances in noncoding RNA modifications of gastrointestinal cancer.Cancer Sci. 2025 Jan;116(1):8-20. doi: 10.1111/cas.16380. Epub 2024 Nov 1. Cancer Sci. 2025. PMID: 39487589 Free PMC article. Review.
-
Structure-guided design of a methyltransferase-like 3 (METTL3) proteolysis targeting chimera (PROTAC) incorporating an indole-nicotinamide chemotype.RSC Med Chem. 2025 Jun 19. doi: 10.1039/d5md00359h. Online ahead of print. RSC Med Chem. 2025. PMID: 40599585 Free PMC article.
-
Targeting METTL3 protein by proteolysis-targeting chimeras: A novel therapeutic approach for acute myeloid leukemia.Genes Dis. 2024 Nov 7;12(4):101452. doi: 10.1016/j.gendis.2024.101452. eCollection 2025 Jul. Genes Dis. 2024. PMID: 40453361 Free PMC article.
References
-
- Boccaletto P.; Stefaniak F.; Ray A.; Cappannini A.; Mukherjee S.; Purta E.; Kurkowska M.; Shirvanizadeh N.; Destefanis E.; Groza P.; Avşar G.; Romitelli A.; Pir P.; Dassi E.; Conticello S. G.; Aguilo F.; Bujnicki J. M.. MODOMICS: A Database of RNA Modification Pathways. 2021 Update. Nucleic Acids Res. 2021, 50. 10.1093/nar/gkab1083. - DOI - PMC - PubMed
-
- Dominissini D.; Moshitch-Moshkovitz S.; Schwartz S.; Salmon-Divon M.; Ungar L.; Osenberg S.; Cesarkas K.; Jacob-Hirsch J.; Amariglio N.; Kupiec M.; Sorek R.; Rechavi G.. Topology of the Human and Mouse m6A RNA Methylomes Revealed by m6A-Seq. 2012, 485 ( (7397), ), 201–206. 10.1038/nature11112. - DOI - PubMed
LinkOut - more resources
Full Text Sources
