Exploring the Landscape of Aptamers: From Cross-Reactive to Selective to Specific, High-Affinity Receptors for Cocaine
- PMID: 38425914
- PMCID: PMC10900216
- DOI: 10.1021/jacsau.3c00781
Exploring the Landscape of Aptamers: From Cross-Reactive to Selective to Specific, High-Affinity Receptors for Cocaine
Abstract
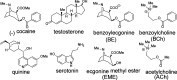
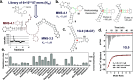
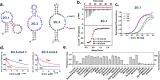
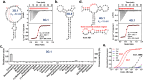
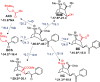
We reported over 20 years ago MNS-4.1, the first DNA aptamer with a micromolar affinity for cocaine. MNS-4.1 is based on a structural motif that is very common in any random pool of oligonucleotides, and it is actually a nonspecific hydrophobic receptor with wide cross-reactivity with alkaloids and steroids. Despite such weaknesses preventing broad applications, this aptamer became widely used in proof-of-concept demonstrations of new formats of biosensors. We now report a series of progressively improved DNA aptamers recognizing cocaine, with the final optimized receptors having low nanomolar affinity and over a thousand-fold selectivity over the initial cross-reactants. In the process of optimization, we tested different methods to eliminate cross-reactivities and improve affinity, eventually achieving properties that are comparable to those of the reported monoclonal antibody candidates for the therapy of overdose. Multiple aptamers that we now report share structural motifs with the previously reported receptor for serotonin. Further mutagenesis studies revealed a palindromic, highly adaptable, broadly cross-reactive hydrophobic motif that could be rebuilt through mutagenesis, expansion of linker regions, and selections into receptors with exceptional affinities and varying specificities.
© 2024 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
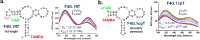
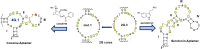
Similar articles
-
Advancements in Aptamer Discovery Technologies.Acc Chem Res. 2016 Sep 20;49(9):1903-10. doi: 10.1021/acs.accounts.6b00283. Epub 2016 Aug 15. Acc Chem Res. 2016. PMID: 27526193
-
Pushing Adenosine and ATP SELEX for DNA Aptamers with Nanomolar Affinity.J Am Chem Soc. 2023 Apr 5;145(13):7540-7547. doi: 10.1021/jacs.3c00848. Epub 2023 Mar 22. J Am Chem Soc. 2023. PMID: 36947745
-
RAID3--An interleukin-6 receptor-binding aptamer with post-selective modification-resistant affinity.RNA Biol. 2015;12(9):1043-53. doi: 10.1080/15476286.2015.1079681. RNA Biol. 2015. PMID: 26383776 Free PMC article.
-
Methods for Improving Aptamer Binding Affinity.Molecules. 2016 Mar 28;21(4):421. doi: 10.3390/molecules21040421. Molecules. 2016. PMID: 27043498 Free PMC article. Review.
-
Nanomaterial-based cocaine aptasensors.Biosens Bioelectron. 2015 Jun 15;68:95-106. doi: 10.1016/j.bios.2014.12.052. Epub 2014 Dec 24. Biosens Bioelectron. 2015. PMID: 25562736 Review.
Cited by
-
An Ultrahigh Affinity DNA Aptamer for Quinine and Its Intrinsic Fluorescence Based Label-Free Detection.Chemistry. 2025 Jan 2;31(1):e202403435. doi: 10.1002/chem.202403435. Epub 2024 Nov 19. Chemistry. 2025. PMID: 39441702 Free PMC article.
-
Examining the Relationship between Aptamer Complexity and Molecular Discrimination of a Low-Epitope Target.ACS Cent Sci. 2024 Nov 11;10(12):2213-2228. doi: 10.1021/acscentsci.4c01377. eCollection 2024 Dec 25. ACS Cent Sci. 2024. PMID: 39735321 Free PMC article.
-
Exploring the relationship between aptamer binding thermodynamics, affinity, and specificity.Nucleic Acids Res. 2025 Mar 20;53(6):gkaf219. doi: 10.1093/nar/gkaf219. Nucleic Acids Res. 2025. PMID: 40156861 Free PMC article.
References
-
- Dunn M. R.; Jimenez R. M.; Chaput J. C. Analysis of aptamer discovery and technology. Nat. Rev. Chem. 2017, 1, 0076.10.1038/s41570-017-0076. - DOI
LinkOut - more resources
Full Text Sources
