Development of a high-density sub-species-specific targeted SNP assay for Rocky Mountain bighorn sheep (Ovis canadensis canadensis)
- PMID: 38426129
- PMCID: PMC10903336
- DOI: 10.7717/peerj.16946
Development of a high-density sub-species-specific targeted SNP assay for Rocky Mountain bighorn sheep (Ovis canadensis canadensis)
Abstract
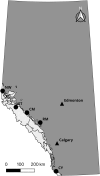
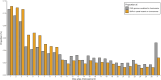
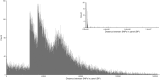
Due to their abundance and relative ease of genotyping, single nucleotide polymorphisms (SNPs) are a commonly used molecular marker for contemporary population genetic and genomic studies. A high-density and cost-effective way to type SNP loci is Allegro targeted genotyping (ATG), which is a form of targeted genotyping by sequencing developed and offered by Tecan genomics. One major drawback of this technology is the need for a reference genome and information on SNP loci when designing a SNP assay. However, for some non-model species genomic information from other closely related species can be used. Here we describe our process of developing an ATG assay to target 50,000 SNPs in Rocky Mountain bighorn sheep, using a reference genome from domestic sheep and SNP resources from prior bighorn sheep studies. We successfully developed a high accuracy, high-density, and relatively low-cost SNP assay for genotyping Rocky Mountain bighorn sheep that genotyped ~45,000 SNP loci. These loci were relatively evenly distributed throughout the genome. Furthermore, the assay produced genotypes at tens of thousands of SNP loci when tested on other mountain sheep species and subspecies.
Keywords: Allegro; Bighorn; Development; Genetic; Genomic; SNP; SPET; Sheep; Tecan; Ungulate.
© 2024 Deakin and Coltman.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




Similar articles
-
Harnessing cross-species alignment to discover SNPs and generate a draft genome sequence of a bighorn sheep (Ovis canadensis).BMC Genomics. 2015 May 20;16(1):397. doi: 10.1186/s12864-015-1618-x. BMC Genomics. 2015. PMID: 25990117 Free PMC article.
-
Whole-genome resequencing uncovers molecular signatures of natural and sexual selection in wild bighorn sheep.Mol Ecol. 2015 Nov;24(22):5616-32. doi: 10.1111/mec.13415. Epub 2015 Nov 6. Mol Ecol. 2015. PMID: 26454263
-
A bioinformatic pipeline for identifying informative SNP panels for parentage assignment from RADseq data.Mol Ecol Resour. 2018 Nov;18(6):1263-1281. doi: 10.1111/1755-0998.12910. Epub 2018 Jul 9. Mol Ecol Resour. 2018. PMID: 29870119 Free PMC article.
-
Genetic linkage map of a wild genome: genomic structure, recombination and sexual dimorphism in bighorn sheep.BMC Genomics. 2010 Sep 28;11:524. doi: 10.1186/1471-2164-11-524. BMC Genomics. 2010. PMID: 20920197 Free PMC article.
-
Recent advances in the genomic resources for sheep.Mamm Genome. 2023 Dec;34(4):545-558. doi: 10.1007/s00335-023-10018-z. Epub 2023 Sep 26. Mamm Genome. 2023. PMID: 37752302 Free PMC article. Review.
References
-
- Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
-
- Andrews KR, Hunter SS, Torrevillas BK, Céspedes N, Garrison SM, Strickland J, Wagers D, Hansten G, New DD, Fagnan MW. A new mouse SNP genotyping assay for speed congenics: combining flexibility, affordability, and power. BMC Genomics. 2021;22:378. doi: 10.1186/s12864-021-07698-9. - DOI - PMC - PubMed
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources

