Identification and functional analysis of ovarian lncRNAs during different egg laying periods in Taihe Black-Bone Chickens
- PMID: 38426211
- PMCID: PMC10902129
- DOI: 10.3389/fphys.2024.1358682
Identification and functional analysis of ovarian lncRNAs during different egg laying periods in Taihe Black-Bone Chickens
Abstract
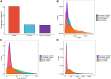
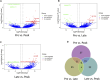
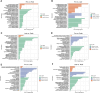
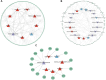
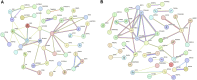
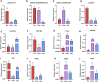
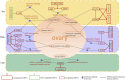
Introduction: Long non-coding RNA (lncRNA) refers to a category of non-coding RNA molecules exceeding 200 nucleotides in length, which exerts a regulatory role in the context of ovarian development. There is a paucity of research examining the involvement of lncRNA in the regulation of ovary development in Taihe Black-Bone Chickens. In order to further investigate the egg laying regulation mechanisms of Taihe Black-Bone Chickens at different periods, transcriptome analysis was conducted on the ovarian tissues at different laying periods. Methods: This study randomly selected ovarian tissues from 12 chickens for RNA-seq. Four chickens were selected for each period, including the early laying period (102 days, Pre), the peak laying period (203 days, Peak), and the late laying period (394 days, Late). Based on our previous study of mRNA expression profiles in the same ovarian tissue, we identified three differentially expressed lncRNAs (DE lncRNAs) at different periods and searched for their cis- and trans-target genes to draw an lncRNA-mRNA network. Results and discussion: In three groups of ovarian tissues, we identified 136 DE lncRNAs, with 8 showing specific expression during the early laying period, 10 showing specific expression during the peak laying period, and 4 showing specific expression during the late laying period. The lncRNA-mRNA network revealed 16 pairs of lncRNA-target genes associated with 7 DE lncRNAs, and these 14 target genes were involved in the regulation of reproductive traits. Furthermore, these reproductive-related target genes were primarily associated with signaling pathways related to follicle and ovary development in Taihe Black-Bone Chickens, including cytokine-cytokine receptor interaction, TGF-beta signaling pathway, tyrosine metabolism, ECM-receptor interaction, focal adhesion, neuroactive ligand-receptor interaction, and cell adhesion molecules (CAMs). This study offers valuable insights for a comprehensive understanding of the influence of lncRNAs on poultry reproductive traits.
Keywords: RNA-seq; Taihe Black-Bone Chicken; egg production; long non-coding RNA; ovary.
Copyright © 2024 Huang, Li, Tan, Xu, Huang and Yin.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Chen C. F., Shiue Y. L., Yen C. J., Tang P. C., Chang H. C., Lee Y. P. (2007). Laying traits and underlying transcripts, expressed in the hypothalamus and pituitary gland, that were associated with egg production variability in chickens. Theriogenology 68 (9), 1305–1315. 10.1016/j.theriogenology.2007.08.032 - DOI - PubMed
-
- Chomchuen K., Tuntiyasawasdikul V., Chankitisakul V., Boonkum W. (2022). Genetic evaluation of body weights and egg production traits using a multi-trait animal model and selection Index in Thai native synthetic chickens (kaimook e-san2). Anim. (Basel) 12 (3), 335. 10.3390/ani12030335 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

