Conserved structures and dynamics in 5'-proximal regions of Betacoronavirus RNA genomes
- PMID: 38426934
- PMCID: PMC11014237
- DOI: 10.1093/nar/gkae144
Conserved structures and dynamics in 5'-proximal regions of Betacoronavirus RNA genomes
Abstract
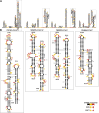
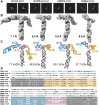
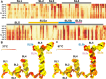
Betacoronaviruses are a genus within the Coronaviridae family of RNA viruses. They are capable of infecting vertebrates and causing epidemics as well as global pandemics in humans. Mitigating the threat posed by Betacoronaviruses requires an understanding of their molecular diversity. The development of novel antivirals hinges on understanding the key regulatory elements within the viral RNA genomes, in particular the 5'-proximal region, which is pivotal for viral protein synthesis. Using a combination of cryo-electron microscopy, atomic force microscopy, chemical probing, and computational modeling, we determined the structures of 5'-proximal regions in RNA genomes of Betacoronaviruses from four subgenera: OC43-CoV, SARS-CoV-2, MERS-CoV, and Rousettus bat-CoV. We obtained cryo-electron microscopy maps and determined atomic-resolution models for the stem-loop-5 (SL5) region at the translation start site and found that despite low sequence similarity and variable length of the helical elements it exhibits a remarkable structural conservation. Atomic force microscopy imaging revealed a common domain organization and a dynamic arrangement of structural elements connected with flexible linkers across all four Betacoronavirus subgenera. Together, these results reveal common features of a critical regulatory region shared between different Betacoronavirus RNA genomes, which may allow targeting of these RNAs by broad-spectrum antiviral therapeutics.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
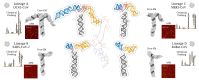


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

