Structure of the PCNA unloader Elg1-RFC
- PMID: 38427736
- PMCID: PMC10906927
- DOI: 10.1126/sciadv.adl1739
Structure of the PCNA unloader Elg1-RFC
Abstract
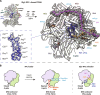
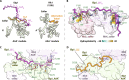
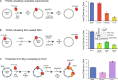
During DNA replication, the proliferating cell nuclear antigen (PCNA) clamps are loaded onto primed sites for each Okazaki fragment synthesis by the AAA+ heteropentamer replication factor C (RFC). PCNA encircling duplex DNA is quite stable and is removed from DNA by the dedicated clamp unloader Elg1-RFC. Here, we show the cryo-EM structure of Elg1-RFC in various states with PCNA. The structures reveal essential features of Elg1-RFC that explain how it is dedicated to PCNA unloading. Specifically, Elg1 contains two external loops that block opening of the Elg1-RFC complex for DNA binding, and an "Elg1 plug" domain that fills the central DNA binding chamber, thereby reinforcing the exclusive PCNA unloading activity of Elg1-RFC. Elg1-RFC was capable of unloading PCNA using non-hydrolyzable AMP-PNP. Both RFC and Elg1-RFC could remove PCNA from covalently closed circular DNA, indicating that PCNA unloading occurs by a mechanism that is distinct from PCNA loading. Implications for the PCNA unloading mechanism are discussed.
Figures



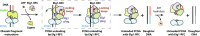
References
-
- A. Kornberg, T. A. Baker, DNA Replication (W.H. Freeman, 1992).
-
- Kastan M. B., DNA damage responses: Mechanisms and roles in human disease: 2007 G.H.A. Clowes Memorial Award Lecture. Mol. Cancer Res. 6, 517–524 (2008). - PubMed
-
- Kong X. P., Onrust R., O'Donnell M., Kuriyan J., Three-dimensional structure of the beta subunit of E. coli DNA polymerase III holoenzyme: A sliding DNA clamp. Cell 69, 425–437 (1992). - PubMed
-
- Gulbis J. M., Kelman Z., Hurwitz J., O'Donnell M., Kuriyan J., Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell 87, 297–306 (1996). - PubMed
-
- Krishna T. S., Kong X. P., Gary S., Burgers P. M., Kuriyan J., Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell 79, 1233–1243 (1994). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

