Real-Time Biosynthetic Reaction Monitoring Informs the Mechanism of Action of Antibiotics
- PMID: 38428018
- PMCID: PMC10941186
- DOI: 10.1021/jacs.4c00081
Real-Time Biosynthetic Reaction Monitoring Informs the Mechanism of Action of Antibiotics
Abstract
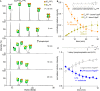
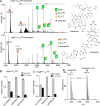
The rapid spread of drug-resistant pathogens and the declining discovery of new antibiotics have created a global health crisis and heightened interest in the search for novel antibiotics. Beyond their discovery, elucidating mechanisms of action has necessitated new approaches, especially for antibiotics that interact with lipidic substrates and membrane proteins. Here, we develop a methodology for real-time reaction monitoring of the activities of two bacterial membrane phosphatases, UppP and PgpB. We then show how we can inhibit their activities using existing and newly discovered antibiotics such as bacitracin and teixobactin. Additionally, we found that the UppP dimer is stabilized by phosphatidylethanolamine, which, unexpectedly, enhanced the speed of substrate processing. Overall, our results demonstrate the potential of native mass spectrometry for real-time biosynthetic reaction monitoring of membrane enzymes, as well as their in situ inhibition and cofactor binding, to inform the mode of action of emerging antibiotics.
Conflict of interest statement
The authors declare the following competing financial interest(s): C.V.R. is a cofounder of and consultant at OMass Therapeutics. The remaining authors declare no competing interests.
Figures



References
-
- Chu J.; Vila-Farres X.; Inoyama D.; Ternei M.; Cohen L. J.; Gordon E. A.; Reddy B. V. B.; Charlop-Powers Z.; Zebroski H. A.; Gallardo-Macias R.; et al. Discovery of MRSA active antibiotics using primary sequence from the human microbiome. Nat. Chem. Biol. 2016, 12 (12), 1004–1006. 10.1038/nchembio.2207. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

