A cryptic plasmid is among the most numerous genetic elements in the human gut
- PMID: 38428395
- PMCID: PMC10973873
- DOI: 10.1016/j.cell.2024.01.039
A cryptic plasmid is among the most numerous genetic elements in the human gut
Abstract
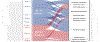
Plasmids are extrachromosomal genetic elements that often encode fitness-enhancing features. However, many bacteria carry "cryptic" plasmids that do not confer clear beneficial functions. We identified one such cryptic plasmid, pBI143, which is ubiquitous across industrialized gut microbiomes and is 14 times as numerous as crAssphage, currently established as the most abundant extrachromosomal genetic element in the human gut. The majority of mutations in pBI143 accumulate in specific positions across thousands of metagenomes, indicating strong purifying selection. pBI143 is monoclonal in most individuals, likely due to the priority effect of the version first acquired, often from one's mother. pBI143 can transfer between Bacteroidales, and although it does not appear to impact bacterial host fitness in vivo, it can transiently acquire additional genetic content. We identified important practical applications of pBI143, including its use in identifying human fecal contamination and its potential as an alternative approach to track human colonic inflammatory states.
Keywords: Bacteroides; cryptic; horizontal gene transfer; human gut microbiome; inflammatory bowel disease; metagenomics; mobile genetic element; plasmid; sewage.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


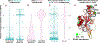

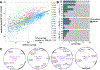
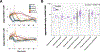
Update of
-
A highly conserved and globally prevalent cryptic plasmid is among the most numerous mobile genetic elements in the human gut.bioRxiv [Preprint]. 2023 Mar 25:2023.03.25.534219. doi: 10.1101/2023.03.25.534219. bioRxiv. 2023. Update in: Cell. 2024 Feb 29;187(5):1206-1222.e16. doi: 10.1016/j.cell.2024.01.039. PMID: 36993556 Free PMC article. Updated. Preprint.
References
-
- Frost LS, Leplae R, Summers AO, and Toussaint A (2005). Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 3, 722–732. - PubMed
-
- Black BE (2017). Centromeres and Kinetochores: Discovering the Molecular Mechanisms Underlying Chromosome Inheritance (Springer; ).
-
- Garoña A, and Dagan T (2021). Darwinian individuality of extrachromosomal genetic elements calls for population genetics tinkering. Environ. Microbiol. Rep. 13, 22–26. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

