Mediator complex subunit 1 architects a tumorigenic Treg cell program independent of inflammation
- PMID: 38428427
- PMCID: PMC10983042
- DOI: 10.1016/j.xcrm.2024.101441
Mediator complex subunit 1 architects a tumorigenic Treg cell program independent of inflammation
Abstract
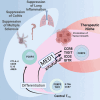
While immunotherapy has revolutionized cancer treatment, its safety has been hampered by immunotherapy-related adverse events. Unexpectedly, we show that Mediator complex subunit 1 (MED1) is required for T regulatory (Treg) cell function specifically in the tumor microenvironment. Treg cell-specific MED1 deletion does not predispose mice to autoimmunity or excessive inflammation. In contrast, MED1 is required for Treg cell promotion of tumor growth because MED1 is required for the terminal differentiation of effector Treg cells in the tumor. Suppression of these terminally differentiated Treg cells is sufficient for eliciting antitumor immunity. Both human and murine Treg cells experience divergent paths of differentiation in tumors and matched tissues with non-malignant inflammation. Collectively, we identify a pathway promoting the differentiation of a Treg cell effector subset specific to tumors and demonstrate that suppression of a subset of Treg cells is sufficient for promoting antitumor immunity in the absence of autoimmune consequences.
Keywords: ATAC-seq; FOXP3; MED1; autoimmunity; differentiation; scRNA-seq; tumor T regulatory cell; tumor immunology.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests B.D.S. holds United States Patent No. US 10,905,706 B2, “Compositions and Methods to Accelerate Resolution of Acute Lung Inflammation” and serves on the Scientific Advisory Board of Zoe Biosciences, outside of the submitted work.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

