Caspase-2 protects against ferroptotic cell death
- PMID: 38429264
- PMCID: PMC10907636
- DOI: 10.1038/s41419-024-06560-6
Caspase-2 protects against ferroptotic cell death
Abstract
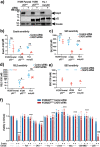
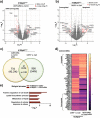
Caspase-2, one of the most evolutionarily conserved members of the caspase family, is an important regulator of the cellular response to oxidative stress. Given that ferroptosis is suppressed by antioxidant defense pathways, such as that involving selenoenzyme glutathione peroxidase 4 (GPX4), we hypothesized that caspase-2 may play a role in regulating ferroptosis. This study provides the first demonstration of an important and unprecedented function of caspase-2 in protecting cancer cells from undergoing ferroptotic cell death. Specifically, we show that depletion of caspase-2 leads to the downregulation of stress response genes including SESN2, HMOX1, SLC7A11, and sensitizes mutant-p53 cancer cells to cell death induced by various ferroptosis-inducing compounds. Importantly, the canonical catalytic activity of caspase-2 is not required for its role and suggests that caspase-2 regulates ferroptosis via non-proteolytic interaction with other proteins. Using an unbiased BioID proteomics screen, we identified novel caspase-2 interacting proteins (including heat shock proteins and co-chaperones) that regulate cellular responses to stress. Finally, we demonstrate that caspase-2 limits chaperone-mediated autophagic degradation of GPX4 to promote the survival of mutant-p53 cancer cells. In conclusion, we document a novel role for caspase-2 as a negative regulator of ferroptosis in cells with mutant p53. Our results provide evidence for a novel function of caspase-2 in cell death regulation and open potential new avenues to exploit ferroptosis in cancer therapy.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Dorstyn L, Hackett-Jones E, Nikolic A, Norris MD, Lim Y, Toubia J, et al. Transcriptome profiling of caspase-2 deficient EmuMyc and Th-MYCN mouse tumors identifies distinct putative roles for caspase-2 in neuronal differentiation and immune signaling. Cell Death Dis. 2019;10:56. doi: 10.1038/s41419-018-1296-0. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

