Predictive role of ctDNA in esophageal squamous cell carcinoma receiving definitive chemoradiotherapy combined with toripalimab
- PMID: 38429311
- PMCID: PMC10907344
- DOI: 10.1038/s41467-024-46307-7
Predictive role of ctDNA in esophageal squamous cell carcinoma receiving definitive chemoradiotherapy combined with toripalimab
Abstract
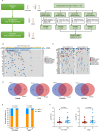
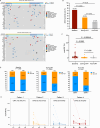
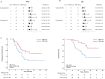
The combination of toripalimab (an anti-PD-1 antibody) with definitive chemoradiotherapy (CRT) demonstrated encouraging efficacy against locally advanced esophageal squamous cell carcinoma (ESCC) in the EC-CRT-001 phase II trial (NCT04005170). The primary endpoint of this trial was the clinical complete response rate (cCR), and the secondary endpoints included overall survival (OS), progression-free survival (PFS), duration of response, and quality of life. The exploratory analyses of EC-CRT-001 include exploring the role of circulating tumor DNA (ctDNA) and blood-based tumor mutational burden (bTMB) in predicting the response and survival. In total, 118 blood and 35 tissue samples from 42 enrolled patients were included in the analyses. We found that ctDNA-negative patients achieved a higher cCR compared to those with detectable ctDNA during CRT (83%, 19/23 vs. 39%, 7/18; p = 0.008) or post-CRT (78%, 21/27 vs. 30%, 3/10; p = 0.017). Patients with detectable ctDNA during CRT had shorter PFS (p = 0.014). Similarly, patients with post-CRT detectable ctDNA had a significantly shorter PFS (p = 0.012) and worse OS (p = 0.004). Moreover, patients with high bTMB levels during CRT had prolonged OS (p = 0.027). In conclusion, ctDNA and bTMB have the potential to predict treatment efficacy and survival in ESCC treated with CRT and immunotherapy.
© 2024. The Author(s).
Conflict of interest statement
Y.Z. is the employee of Nanjing Geneseeq Technology Inc. All other authors have no conflicts of interest to declare.
Figures


References
-
- Luo H, et al. Effect of Camrelizumab vs Placebo Added to Chemotherapy on Survival and Progression-Free Survival in Patients With Advanced or Metastatic Esophageal Squamous Cell Carcinoma: The ESCORT-1st Randomized Clinical Trial. JAMA. 2021;326:916–925. doi: 10.1001/jama.2021.12836. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

