G2019S selective LRRK2 kinase inhibitor abrogates mitochondrial DNA damage
- PMID: 38429321
- PMCID: PMC10907374
- DOI: 10.1038/s41531-024-00660-y
G2019S selective LRRK2 kinase inhibitor abrogates mitochondrial DNA damage
Abstract
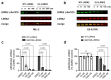
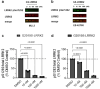
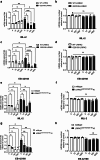
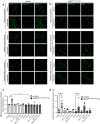
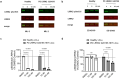
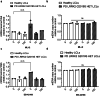
Pathogenic mutations in LRRK2 cause Parkinson's disease (PD). The G2019S variant is the most common, which results in abnormally high kinase activity. Compounds that target LRRK2 kinase activity are currently being developed and tested in clinical trials. We recently found that G2019S LRRK2 causes mitochondrial DNA (mtDNA) damage and treatment with multiple classes of LRRK2 kinase inhibitors at concentrations associated with dephosphorylation of LRRK2 reversed mtDNA damage to healthy control levels. Because maintaining the normal function of LRRK2 in heterozygous G2019S LRRK2 carriers while specifically targeting the G2019S LRRK2 activity could have an advantageous safety profile, we explored the efficacy of a G2019S mutant selective LRRK2 inhibitor to reverse mtDNA damage in G2019S LRRK2 models and patient cells relative to non-selective LRRK2 inhibitors. Potency of LRRK2 kinase inhibition by EB-42168, a G2019S mutant LRRK2 kinase inhibitor, and MLi-2, a non-selective inhibitor, was determined by measuring phosphorylation of LRRK2 at Ser935 and/or Ser1292 using quantitative western immunoblot analysis. The Mito DNADX assay, which allows for the accurate real-time quantification of mtDNA damage in a 96-well platform, was performed in parallel. We confirmed that EB-42168 selectively inhibits LRRK2 phosphorylation on G2019S LRRK2 relative to wild-type LRRK2. On the other hand, MLi-2 was equipotent for wild-type and G2019S LRRK2. Acute treatment with EB-42168 inhibited LRRK2 phosphorylation and also restored mtDNA damage to healthy control levels. We further investigated the relationship between LRRK2 kinase activity, mtDNA damage and mitophagy. Levels of mtDNA damage caused by G2019S LRRK2 were fully re-established within 2 h of a LRRK2 inhibitor wash out and recovery experiment, indicating the mtDNA damage phenotype is highly dynamic. G2019S LRRK2 mitophagy defects were not alleviated with LRRK2 kinase inhibition, suggesting that mitophagy is not mechanistically regulating LRRK2 kinase-mediated reversal of mtDNA damage in this acute timeframe. Abrogation of mtDNA damage with the mutant selective tool inhibitor EB-42168 demonstrates the potential of a precision medicine approach for LRRK2 G2019S PD. Levels of mtDNA damage may serve as a potential pharmacodynamic biomarker of altered kinase activity that could be useful for small molecule development and clinical trials.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests. The former employees of Escape Bio do not have any financial ties to what is now a former company. They are all now employed by different companies and are committed to seeing the work made available to the scientific community and do not have any conflicts of interest as stated.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

