The first mitochondrial genome of Calophyllum soulattri Burm.f
- PMID: 38429360
- PMCID: PMC10907642
- DOI: 10.1038/s41598-024-55016-6
The first mitochondrial genome of Calophyllum soulattri Burm.f
Abstract
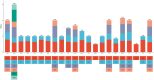
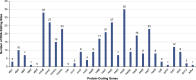
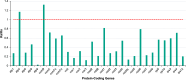
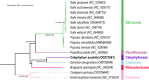
Calophyllum soulattri Burm.f. is traditionally used to treat skin infections and reduce rheumatic pain, yet genetic and genomic studies are still limited. Here, we present the first complete mitochondrial genome of C. soulattri. It is 378,262 bp long with 43.97% GC content, containing 55 genes (30 protein-coding, 5 rRNA, and 20 tRNA). Repeat analysis of the mitochondrial genome revealed 194 SSRs, mostly mononucleotides, and 266 pairs of dispersed repeats ( 30 bp) that were predominantly palindromic. There were 23 homologous fragments found between the mitochondrial and plastome genomes. We also predicted 345 C-to-U RNA editing sites from 30 protein-coding genes (PCGs) of the C. soulatrii mitochondrial genome. These RNA editing events created the start codon of nad1 and the stop codon of ccmFc. Most PCGs of the C. soulattri mitochondrial genome underwent negative selection, but atp4 and ccmB experienced positive selection. Phylogenetic analyses showed C. soulattri is a sister taxon of Garcinia mangostana. This study has shed light on C. soulattri's evolution and Malpighiales' phylogeny. As the first complete mitochondrial genome in Calophyllaceae, it can be used as a reference genome for other medicinal plant species within the family for future genetic studies.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
The complete chloroplast genome of Calophyllum soulattri Burm. f. (Calophyllaceae).Mitochondrial DNA B Resour. 2023 May 25;8(5):607-611. doi: 10.1080/23802359.2023.2215350. eCollection 2023. Mitochondrial DNA B Resour. 2023. PMID: 37250208 Free PMC article.
-
Comparative mitochondrial genomics of Terniopsis yongtaiensis in Malpighiales: structural, sequential, and phylogenetic perspectives.BMC Genomics. 2024 Sep 12;25(1):853. doi: 10.1186/s12864-024-10765-6. BMC Genomics. 2024. PMID: 39267005 Free PMC article.
-
Assembly and characterization of the first mitochondrial genome of Phyllanthaceae: a case study of the ornamental aquatic plant Phyllanthus fluitans.Genetica. 2025 Jul 16;153(1):25. doi: 10.1007/s10709-025-00241-8. Genetica. 2025. PMID: 40668455
-
Assembly and comparative analysis of the first complete mitochondrial genome of Astragalus membranaceus (Fisch.) Bunge: an invaluable traditional Chinese medicine.BMC Plant Biol. 2024 Nov 8;24(1):1055. doi: 10.1186/s12870-024-05780-4. BMC Plant Biol. 2024. PMID: 39511474 Free PMC article.
-
Mitochondrial genome of Garcinia mangostana L. variety Mesta.Sci Rep. 2022 Jun 8;12(1):9480. doi: 10.1038/s41598-022-13706-z. Sci Rep. 2022. PMID: 35676406 Free PMC article.
Cited by
-
Comparative analysis of the mitogenomes of multiple species of Fagaceae, with special focus on Quercus gilva.BMC Plant Biol. 2025 Aug 19;25(1):1098. doi: 10.1186/s12870-025-07072-x. BMC Plant Biol. 2025. PMID: 40830429 Free PMC article.
-
Characterization of the complete chloroplast genome of Garcinia xanthochymus Hook. f. ex T. Anderson 1874 (Clusiaceae) and its phylogenetic implications.Mitochondrial DNA B Resour. 2025 Feb 21;10(3):262-266. doi: 10.1080/23802359.2025.2468760. eCollection 2025. Mitochondrial DNA B Resour. 2025. PMID: 39995956 Free PMC article.
-
Assembly and analysis of the complete mitochondrial and chloroplast genomes of Vigna reflexo-pilosa.PLoS One. 2025 Jun 11;20(6):e0325243. doi: 10.1371/journal.pone.0325243. eCollection 2025. PLoS One. 2025. PMID: 40498740 Free PMC article.
References
-
- Group et al. An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: Apg IV. Bot. J. Linnean Soc.181, 1–20 (2016).
-
- Gupta, S. & Gupta, P. The genus Calophyllum: review of ethnomedicinal uses, phytochemistry and pharmacology. Bioactive Natural Prod. Drug Discovery 215–242 (2020).
-
- Lim, C.-K., Hemaroopini, S., Say, Y.-H. & Jong, V. Y.-M. Cytotoxic compounds from the stem bark of Calophyllum soulattri. Natural Product Commun.12, 1934578X1701200922 (2017).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

