Sarcoma microenvironment cell states and ecosystems are associated with prognosis and predict response to immunotherapy
- PMID: 38429415
- PMCID: PMC11058033
- DOI: 10.1038/s43018-024-00743-y
Sarcoma microenvironment cell states and ecosystems are associated with prognosis and predict response to immunotherapy
Abstract
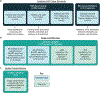
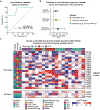
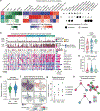
Characterization of the diverse malignant and stromal cell states that make up soft tissue sarcomas and their correlation with patient outcomes has proven difficult using fixed clinical specimens. Here, we employed EcoTyper, a machine-learning framework, to identify the fundamental cell states and cellular ecosystems that make up sarcomas on a large scale using bulk transcriptomes with clinical annotations. We identified and validated 23 sarcoma-specific, transcriptionally defined cell states, many of which were highly prognostic of patient outcomes across independent datasets. We discovered three conserved cellular communities or ecotypes associated with underlying genomic alterations and distinct clinical outcomes. We show that one ecotype defined by tumor-associated macrophages and epithelial-like malignant cells predicts response to immune-checkpoint inhibition but not chemotherapy and validate our findings in an independent cohort. Our results may enable identification of patients with soft tissue sarcomas who could benefit from immunotherapy and help develop new therapeutic strategies.
© 2024. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
COMPETING INTERESTS STATEMENT
W.D.T. has served as a consultant/advisor for Eli Lilly, Daiichi Sankyo, Deciphera, Foghorn Therapeutics, AmMAx Bio, Novo Holdings, Servier, Medpacto, Ayala Pharmaceuticals, Kowa Research Inst., Epizyme Inc (Nexus Global Group), Bayer, Cogent Biosciences, Amgen, and PER, has received research funding from Novartis, Eli Lilly, Plexxikon, Daiichi Sankyo, Tracon Pharma, Blueprint Medicines, Immune Design, BioAlta, and Deciphera, and has a patent on companion diagnostics for CDK4 inhibitors (4/854,329). S.P.D. has served as a consultant/advisor for EMD Serono, Amgen, Nektar, Immune Design, GlaxoSmithKline, Incyte, Merck, Adaptimmune, Immunocore, Pfizer, Servier, Rain Therapeutics, GI Innovations, and Aadi Bioscience and has received research funding from EMD Serono, Amgen, Merck, Incyte, Nektar, Bristol-Meyers Squibb, and Deciphera. E.J.M. has served as a paid consultant for Guidepoint. The other authors declare no competing interests.
Figures



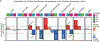


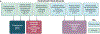





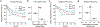

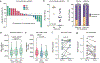
References
-
- Wang D, Zhang Q, Eisenberg BL, Kane JM, Li XA, Lucas D, Petersen IA, DeLaney TF, Freeman CR, Finkelstein SE, et al. (2015). Significant Reduction of Late Toxicities in Patients With Extremity Sarcoma Treated With Image-Guided Radiation Therapy to a Reduced Target Volume: Results of Radiation Therapy Oncology Group RTOG-0630 Trial. J. Clin. Oncol 33, 2231–2238. 10.1200/JCO.2014.58.5828. - DOI - PMC - PubMed
-
- Judson I, Verweij J, Gelderblom H, Hartmann JT, Schöffski P, Blay J-Y, Kerst JM, Sufliarsky J, Whelan J, Hohenberger P, et al. (2014). Doxorubicin alone versus intensified doxorubicin plus ifosfamide for first-line treatment of advanced or metastatic soft-tissue sarcoma: a randomised controlled phase 3 trial. Lancet Oncol. 15, 415–423. 10.1016/S1470-2045(14)70063-4. - DOI - PubMed
-
- Ryan CW, Merimsky O, Agulnik M, Blay J-Y, Schuetze SM, Van Tine BA, Jones RL, Elias AD, Choy E, Alcindor T, et al. (2016). PICASSO III: A Phase III, Placebo-Controlled Study of Doxorubicin With or Without Palifosfamide in Patients With Metastatic Soft Tissue Sarcoma. J Clin Oncol 34, 3898–3905. 10.1200/JCO.2016.67.6684. - DOI - PubMed
-
- Tap WD, Papai Z, Van Tine BA, Attia S, Ganjoo KN, Jones RL, Schuetze S, Reed D, Chawla SP, Riedel RF, et al. (2017). Doxorubicin plus evofosfamide versus doxorubicin alone in locally advanced, unresectable or metastatic soft-tissue sarcoma (TH CR-406/SARC021): an international, multicentre, open-label, randomised phase 3 trial. Lancet Oncol 18, 1089–1103. 10.1016/S1470-2045(17)30381-9. - DOI - PMC - PubMed
Methods-only references
-
- Bui NQ, Nemat-Gorgani N, Subramanian A, Torres IA, Lohman M, Sears TJ, van de Rijn M, Charville GW, Becker H-C, Wang DS, et al. (2023). Monitoring Sarcoma Response to Immune Checkpoint Inhibition and Local Cryotherapy with Circulating Tumor DNA Analysis. Clin Cancer Res 29, 2612–2620. 10.1158/1078-0432.CCR-23-0250. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

