Iron dysregulation and inflammatory stress erythropoiesis associates with long-term outcome of COVID-19
- PMID: 38429458
- PMCID: PMC10907301
- DOI: 10.1038/s41590-024-01754-8
Iron dysregulation and inflammatory stress erythropoiesis associates with long-term outcome of COVID-19
Abstract
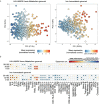
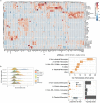
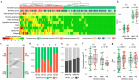
Persistent symptoms following SARS-CoV-2 infection are increasingly reported, although the drivers of post-acute sequelae (PASC) of COVID-19 are unclear. Here we assessed 214 individuals infected with SARS-CoV-2, with varying disease severity, for one year from COVID-19 symptom onset to determine the early correlates of PASC. A multivariate signature detected beyond two weeks of disease, encompassing unresolving inflammation, anemia, low serum iron, altered iron-homeostasis gene expression and emerging stress erythropoiesis; differentiated those who reported PASC months later, irrespective of COVID-19 severity. A whole-blood heme-metabolism signature, enriched in hospitalized patients at month 1-3 post onset, coincided with pronounced iron-deficient reticulocytosis. Lymphopenia and low numbers of dendritic cells persisted in those with PASC, and single-cell analysis reported iron maldistribution, suggesting monocyte iron loading and increased iron demand in proliferating lymphocytes. Thus, defects in iron homeostasis, dysregulated erythropoiesis and immune dysfunction due to COVID-19 possibly contribute to inefficient oxygen transport, inflammatory disequilibrium and persisting symptomatology, and may be therapeutically tractable.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no conflicts of interest.
Figures








References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

