A multi-ancestry genetic study of pain intensity in 598,339 veterans
- PMID: 38429522
- PMCID: PMC12105102
- DOI: 10.1038/s41591-024-02839-5
A multi-ancestry genetic study of pain intensity in 598,339 veterans
Erratum in
-
Author Correction: A multi-ancestry genetic study of pain intensity in 598,339 veterans.Nat Med. 2024 Jul;30(7):2088. doi: 10.1038/s41591-024-03024-4. Nat Med. 2024. PMID: 38714900 No abstract available.
Abstract
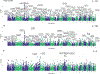
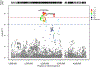
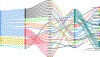
Chronic pain is a common problem, with more than one-fifth of adult Americans reporting pain daily or on most days. It adversely affects the quality of life and imposes substantial personal and economic costs. Efforts to treat chronic pain using opioids had a central role in precipitating the opioid crisis. Despite an estimated heritability of 25-50%, the genetic architecture of chronic pain is not well-characterized, in part because studies have largely been limited to samples of European ancestry. To help address this knowledge gap, we conducted a cross-ancestry meta-analysis of pain intensity in 598,339 participants in the Million Veteran Program, which identified 126 independent genetic loci, 69 of which are new. Pain intensity was genetically correlated with other pain phenotypes, level of substance use and substance use disorders, other psychiatric traits, education level and cognitive traits. Integration of the genome-wide association studies findings with functional genomics data shows enrichment for putatively causal genes (n = 142) and proteins (n = 14) expressed in brain tissues, specifically in GABAergic neurons. Drug repurposing analysis identified anticonvulsants, β-blockers and calcium-channel blockers, among other drug groups, as having potential analgesic effects. Our results provide insights into key molecular contributors to the experience of pain and highlight attractive drug targets.
© 2024. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
H.R.K. is a member of advisory boards for Dicerna Pharmaceuticals, Sophrosyne Pharmaceuticals, Enthion Pharmaceuticals and Clearmind Medicine; a consultant to Sobrera Pharmaceuticals; the recipient of research funding and medication supplies from Alkermes for an investigator-initiated study; and a member of the American Society of Clinical Psychopharmacology’s Alcohol Clinical Trials Initiative, which was supported in the last 3 years by Alkermes, Dicerna, Ethypharm, Lundbeck, Mitsubishi and Otsuka. H.R.K. and J.G. are named as inventors on PCT patent application 15/878,640 entitled ‘genotype-guided dosing of opioid agonists’, filed on 24 January 2018. E.S. is a full-time employee of Regeneron Pharmaceuticals. The other authors declare no competing interests.
Figures

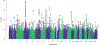

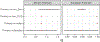


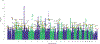
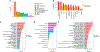
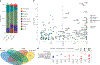

References
-
- Yong RJ, Mullins PM & Bhattacharyya N Prevalence of chronic pain among adults in the United States. Pain 163, e328–e332 (2022). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

