A gut microbial signature for combination immune checkpoint blockade across cancer types
- PMID: 38429524
- PMCID: PMC10957475
- DOI: 10.1038/s41591-024-02823-z
A gut microbial signature for combination immune checkpoint blockade across cancer types
Abstract
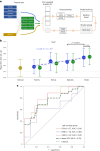
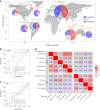
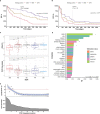
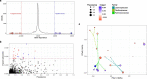
Immune checkpoint blockade (ICB) targeting programmed cell death protein 1 (PD-1) and cytotoxic T lymphocyte protein 4 (CTLA-4) can induce remarkable, yet unpredictable, responses across a variety of cancers. Studies suggest that there is a relationship between a cancer patient's gut microbiota composition and clinical response to ICB; however, defining microbiome-based biomarkers that generalize across cohorts has been challenging. This may relate to previous efforts quantifying microbiota to species (or higher taxonomic rank) abundances, whereas microbial functions are often strain specific. Here, we performed deep shotgun metagenomic sequencing of baseline fecal samples from a unique, richly annotated phase 2 trial cohort of patients with diverse rare cancers treated with combination ICB (n = 106 discovery cohort). We demonstrate that strain-resolved microbial abundances improve machine learning predictions of ICB response and 12-month progression-free survival relative to models built using species-rank quantifications or comprehensive pretreatment clinical factors. Through a meta-analysis of gut metagenomes from a further six comparable studies (n = 364 validation cohort), we found cross-cancer (and cross-country) validity of strain-response signatures, but only when the training and test cohorts used concordant ICB regimens (anti-PD-1 monotherapy or combination anti-PD-1 plus anti-CTLA-4). This suggests that future development of gut microbiome diagnostics or therapeutics should be tailored according to ICB treatment regimen rather than according to cancer type.
© 2024. The Author(s).
Conflict of interest statement
A.G. has received a speaker honorarium from Microbiotica Limited. B.M. has served on advisory boards for Amgen, Bristol Myers Squibb (BMS), Merck, Beigene and AstraZeneca (AZ). M.S.C. has served on advisory boards or as a consultant for Amgen, BMS, Eisai, Ideaya, Merck, Sharp & Dohme (MSD), Nektar, Novartis, Oncosec, Pierre-Fabre, Qbiotics, Regeneron, Roche, Merck, Moderna and Sanofi and received honoraria from BMS, MSD and Novartis. D.K. has served on advisory boards for BMS, MSD and Novartis. C.U. has served in a consulting/advisory role for Merck Serano and AZ and a speakers’ bureau role for IQvia and AZ. His institution has received research funding from Akeso Biopharma, Arcus Biosciences, Atridia, BeyondSpring Pharmaceuticals, Boehringer Ingelheim, Deciphera and Novotech. S.F. has received financial support from Amgen, MSD and AZ; honoraria for advisory boards from Akesobio, Ambrax and MSD; and institutional sponsorship/trials and research activities from Akesobio, Ambrax, Amgen, Axelia, AZ, Aulos, BeiGene, Cullinan, Daiichi Sankyo, Edison Oncology, Genentech, MSD, Takeda, HaiHe Biopharma, Vivace and WellMarker Bio. D.J.A. is a paid consultant for Ono Therapeutics and Microbiotica Limited and receives research support from AZ, OpenTargets and BMS. T.D.L. is cofounder and chief scientific officer at Microbiotica Limited. All other authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

