PHF6-mediated transcriptional control of NSC via Ephrin receptors is impaired in the intellectual disability syndrome BFLS
- PMID: 38429579
- PMCID: PMC10933485
- DOI: 10.1038/s44319-024-00082-0
PHF6-mediated transcriptional control of NSC via Ephrin receptors is impaired in the intellectual disability syndrome BFLS
Abstract
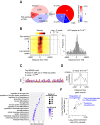
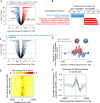
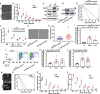
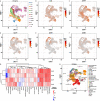
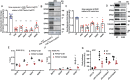
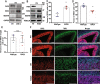
The plant homeodomain zinc-finger protein, PHF6, is a transcriptional regulator, and PHF6 germline mutations cause the X-linked intellectual disability (XLID) Börjeson-Forssman-Lehmann syndrome (BFLS). The mechanisms by which PHF6 regulates transcription and how its mutations cause BFLS remain poorly characterized. Here, we show genome-wide binding of PHF6 in the developing cortex in the vicinity of genes involved in central nervous system development and neurogenesis. Characterization of BFLS mice harbouring PHF6 patient mutations reveals an increase in embryonic neural stem cell (eNSC) self-renewal and a reduction of neural progenitors. We identify a panel of Ephrin receptors (EphRs) as direct transcriptional targets of PHF6. Mechanistically, we show that PHF6 regulation of EphR is impaired in BFLS mice and in conditional Phf6 knock-out mice. Knockdown of EphR-A phenocopies the PHF6 loss-of-function defects in altering eNSCs, and its forced expression rescues defects of BFLS mice-derived eNSCs. Our data indicate that PHF6 directly promotes Ephrin receptor expression to control eNSC behaviour in the developing brain, and that this pathway is impaired in BFLS.
Keywords: BFLS; Ephrin Receptors; Intellectual Disability; Neural Stem Cells; PHF6.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
MeSH terms
Substances
Supplementary concepts
Grants and funding
- RGPIN-2016-00605/Canadian Government | Natural Sciences and Engineering Research Council of Canada (NSERC)
- RGPIN-2018-05962/Canadian Government | Natural Sciences and Engineering Research Council of Canada (NSERC)
- PJG-185800/Canadian HIV Trials Network, Canadian Institutes of Health Research (CTN, CIHR)
- PJT-159619/Canadian HIV Trials Network, Canadian Institutes of Health Research (CTN, CIHR)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

