Cell-free DNA methylation reveals cell-specific tissue injury and correlates with disease severity and patient outcomes in COVID-19
- PMID: 38429730
- PMCID: PMC10908074
- DOI: 10.1186/s13148-024-01645-7
Cell-free DNA methylation reveals cell-specific tissue injury and correlates with disease severity and patient outcomes in COVID-19
Abstract
Background: The recently identified methylation patterns specific to cell type allows the tracing of cell death dynamics at the cellular level in health and diseases. This study used COVID-19 as a disease model to investigate the efficacy of cell-specific cell-free DNA (cfDNA) methylation markers in reflecting or predicting disease severity or outcome.
Methods: Whole genome methylation sequencing of cfDNA was performed for 20 healthy individuals, 20 cases with non-hospitalized COVID-19 and 12 cases with severe COVID-19 admitted to intensive care unit (ICU). Differentially methylated regions (DMRs) and gene ontology pathway enrichment analyses were performed to explore the locus-specific methylation difference between cohorts. The proportion of cfDNA derived from lung and immune cells to a given sample (i.e. tissue fraction) at cell-type resolution was estimated using a novel algorithm, which reflects lung injuries and immune response in COVID-19 patients and was further used to evaluate clinical severity and patient outcome.
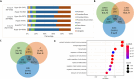
Results: COVID‑19 patients had globally reduced cfDNA methylation level compared with healthy controls. Compared with non-hospitalized COVID-19 patients, the cfDNA methylation pattern was significantly altered in severe patients with the identification of 11,156 DMRs, which were mainly enriched in pathways related to immune response. Markedly elevated levels of cfDNA derived from lung and more specifically alveolar epithelial cells, bronchial epithelial cells, and lung endothelial cells were observed in COVID-19 patients compared with healthy controls. Compared with non-hospitalized patients or healthy controls, severe COVID-19 had significantly higher cfDNA derived from B cells, T cells and granulocytes and lower cfDNA from natural killer cells. Moreover, cfDNA derived from alveolar epithelial cells had the optimal performance to differentiate COVID-19 with different severities, lung injury levels, SOFA scores and in-hospital deaths, with the area under the receiver operating characteristic curve of 0.958, 0.941, 0.919 and 0.955, respectively.
Conclusion: Severe COVID-19 has a distinct cfDNA methylation signature compared with non-hospitalized COVID-19 and healthy controls. Cell type-specific cfDNA methylation signature enables the tracing of COVID-19 related cell deaths in lung and immune cells at cell-type resolution, which is correlated with clinical severities and outcomes, and has extensive application prospects to evaluate tissue injuries in diseases with multi-organ dysfunction.
Keywords: Circulating-free DNA; Coronavirus disease 2019; Immune response; Tissue of origin inference; Whole genome methylation sequencing.
© 2024. The Author(s).
Conflict of interest statement
Ming-Ming Yuan, Jing-Dong Wang, Yu-Fei Wang, Jun Li, Qian Li and Rong-Rong Chen are employees of Geneplus (Beijing, China). The remaining authors report no conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

