Identification of crosstalk genes relating to ECM-receptor interaction genes in MASH and DN using bioinformatics and machine learning
- PMID: 38429902
- PMCID: PMC10907849
- DOI: 10.1111/jcmm.18156
Identification of crosstalk genes relating to ECM-receptor interaction genes in MASH and DN using bioinformatics and machine learning
Abstract
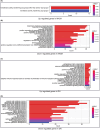
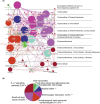
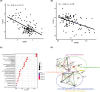
This study aimed to identify genes shared by metabolic dysfunction-associated fatty liver disease (MASH) and diabetic nephropathy (DN) and the effect of extracellular matrix (ECM) receptor interaction genes on them. Datasets with MASH and DN were downloaded from the Gene Expression Omnibus (GEO) database. Pearson's coefficients assessed the correlation between ECM-receptor interaction genes and cross talk genes. The coexpression network of co-expression pairs (CP) genes was integrated with its protein-protein interaction (PPI) network, and machine learning was employed to identify essential disease-representing genes. Finally, immuno-penetration analysis was performed on the MASH and DN gene datasets using the CIBERSORT algorithm to evaluate the plausibility of these genes in diseases. We found 19 key CP genes. Fos proto-oncogene (FOS), belonging to the IL-17 signalling pathway, showed greater centrality PPI network; Hyaluronan Mediated Motility Receptor (HMMR), belonging to ECM-receptor interaction genes, showed most critical in the co-expression network map of 19 CP genes; Forkhead Box C1 (FOXC1), like FOS, showed a high ability to predict disease in XGBoost analysis. Further immune infiltration showed a clear positive correlation between FOS/FOXC1 and mast cells that secrete IL-17 during inflammation. Combining the results of previous studies, we suggest a FOS/FOXC1/HMMR regulatory axis in MASH and DN may be associated with mast cells in the acting IL-17 signalling pathway. Extracellular HMMR may regulate the IL-17 pathway represented by FOS through the Mitogen-Activated Protein Kinase 1 (ERK) or PI3K-Akt-mTOR pathway. HMMR may serve as a signalling carrier between MASH and DN and could be targeted for therapeutic development.
Keywords: DN; ECM-receptor interaction; MASH; XGBoost; immune infiltration.
© 2024 The Authors. Journal of Cellular and Molecular Medicine published by Foundation for Cellular and Molecular Medicine and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures








References
-
- Eslam M, Newsome PN, Sarin SK, et al. A new definition for metabolic dysfunction‐associated fatty liver disease: an international expert consensus statement. J Hepatol. 2020;73:202‐209. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

