Consequences of Amyloid-β Deficiency for the Liver
- PMID: 38430535
- PMCID: PMC11095235
- DOI: 10.1002/advs.202307734
Consequences of Amyloid-β Deficiency for the Liver
Abstract
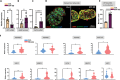
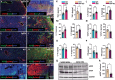
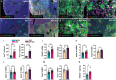
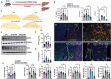
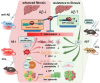
The hepatic content of amyloid beta (Aβ) decreases drastically in human and rodent cirrhosis highlighting the importance of understanding the consequences of Aβ deficiency in the liver. This is especially relevant in view of recent advances in anti-Aβ therapies for Alzheimer's disease (AD). Here, it is shown that partial hepatic loss of Aβ in transgenic AD mice immunized with Aβ antibody 3D6 and its absence in amyloid precursor protein (APP) knockout mice (APP-KO), as well as in human liver spheroids with APP knockdown upregulates classical hallmarks of fibrosis, smooth muscle alpha-actin, and collagen type I. Aβ absence in APP-KO and deficiency in immunized mice lead to strong activation of transforming growth factor-β (TGFβ), alpha secretases, NOTCH pathway, inflammation, decreased permeability of liver sinusoids, and epithelial-mesenchymal transition. Inversely, increased systemic and intrahepatic levels of Aβ42 in transgenic AD mice and neprilysin inhibitor LBQ657-treated wild-type mice protect the liver against carbon tetrachloride (CCl4)-induced injury. Transcriptomic analysis of CCl4-treated transgenic AD mouse livers uncovers the regulatory effects of Aβ42 on mitochondrial function, lipid metabolism, and its onco-suppressive effects accompanied by reduced synthesis of extracellular matrix proteins. Combined, these data reveal Aβ as an indispensable regulator of cell-cell interactions in healthy liver and a powerful protector against liver fibrosis.
Keywords: 5xFAD; TGFβ; VEGF; eNOS; neprilysin; presenilin; β‐secretase 1.
© 2024 The Authors. Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
Eberhard Karls University of Tübingen in conjunction with the University Hospital of Tübingen has filed a patent covering Aβ‐enhancing strategies for the treatment of liver fibrosis where G.H.B, R.W., T.S.W, M.S., and L.D. are listed as inventors. V.M.L is a co‐founder, CEO, and shareholder of HepaPredict AB. All other authors declare no conflict of interest.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
