Biochemical and metabolic characterization of a G6PC2 inhibitor
- PMID: 38431189
- PMCID: PMC11661470
- DOI: 10.1016/j.biochi.2024.02.012
Biochemical and metabolic characterization of a G6PC2 inhibitor
Abstract
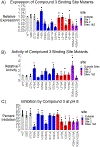
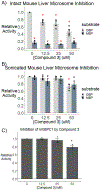
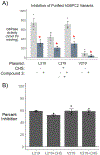
Three glucose-6-phosphatase catalytic subunits, that hydrolyze glucose-6-phosphate (G6P) to glucose and inorganic phosphate, have been identified, designated G6PC1-3, but only G6PC1 and G6PC2 have been implicated in the regulation of fasting blood glucose (FBG). Elevated FBG has been associated with multiple adverse clinical outcomes, including increased risk for type 2 diabetes and various cancers. Therefore, G6PC1 and G6PC2 inhibitors that lower FBG may be of prophylactic value for the prevention of multiple conditions. The studies described here characterize a G6PC2 inhibitor, designated VU0945627, previously identified as Compound 3. We show that VU0945627 preferentially inhibits human G6PC2 versus human G6PC1 but activates human G6PC3. VU0945627 is a mixed G6PC2 inhibitor, increasing the Km but reducing the Vmax for G6P hydrolysis. PyRx virtual docking to an AlphaFold2-derived G6PC2 structural model suggests VU0945627 binds two sites in human G6PC2. Mutation of residues in these sites reduces the inhibitory effect of VU0945627. VU0945627 does not inhibit mouse G6PC2 despite its 84% sequence identity with human G6PC2. Mutagenesis studies suggest this lack of inhibition of mouse G6PC2 is due, in part, to a change in residue 318 from histidine in human G6PC2 to proline in mouse G6PC2. Surprisingly, VU0945627 still inhibited glucose cycling in the mouse islet-derived βTC-3 cell line. Studies using intact mouse liver microsomes and PyRx docking suggest that this observation can be explained by an ability of VU0945627 to also inhibit the G6P transporter SLC37A4. These data will inform future computational modeling studies designed to identify G6PC isoform-specific inhibitors.
Keywords: Enzyme inhibition; Fasting blood glucose; Glucose-6-phosphatase; Islet.
Copyright © 2024 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures






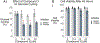

Similar articles
-
Functional Analysis of Mouse G6pc1 Mutations Using a Novel In Situ Assay for Glucose-6-Phosphatase Activity and the Effect of Mutations in Conserved Human G6PC1/G6PC2 Amino Acids on G6PC2 Protein Expression.PLoS One. 2016 Sep 9;11(9):e0162439. doi: 10.1371/journal.pone.0162439. eCollection 2016. PLoS One. 2016. PMID: 27611587 Free PMC article.
-
G6PC2 Modulates the Effects of Dexamethasone on Fasting Blood Glucose and Glucose Tolerance.Endocrinology. 2016 Nov;157(11):4133-4145. doi: 10.1210/en.2016-1678. Epub 2016 Sep 21. Endocrinology. 2016. PMID: 27653037 Free PMC article.
-
Single-incision sling operations for urinary incontinence in women.Cochrane Database Syst Rev. 2017 Jul 26;7(7):CD008709. doi: 10.1002/14651858.CD008709.pub3. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2023 Oct 27;10:CD008709. doi: 10.1002/14651858.CD008709.pub4. PMID: 28746980 Free PMC article. Updated.
-
Prophylactic platelet transfusion for prevention of bleeding in patients with haematological disorders after chemotherapy and stem cell transplantation.Cochrane Database Syst Rev. 2012 May 16;2012(5):CD004269. doi: 10.1002/14651858.CD004269.pub3. Cochrane Database Syst Rev. 2012. PMID: 22592695 Free PMC article.
-
Cost-effectiveness of using prognostic information to select women with breast cancer for adjuvant systemic therapy.Health Technol Assess. 2006 Sep;10(34):iii-iv, ix-xi, 1-204. doi: 10.3310/hta10340. Health Technol Assess. 2006. PMID: 16959170
Cited by
-
Identification and validation of tricarboxylic acid cycle-related diagnostic biomarkers for diabetic nephropathy via weighted gene co-expression network analysis and single-cell transcriptome analysis.Acta Diabetol. 2025 Jul 31. doi: 10.1007/s00592-025-02557-5. Online ahead of print. Acta Diabetol. 2025. PMID: 40742465
References
-
- Arden SD, Zahn T, Steegers S, Webb S, Bergman B, O'Brien RM, and Hutton JC (1999) Molecular cloning of a pancreatic islet-specific glucose-6-phosphatase catalytic subunit-related protein. Diabetes 48, 531–542 - PubMed
-
- Martin CC, Bischof LJ, Bergman B, Hornbuckle LA, Hilliker C, Frigeri C, Wahl D, Svitek CA, Wong R, Goldman JK, Oeser JK, Lepretre F, Froguel P, O'Brien RM, and Hutton JC (2001) Cloning and Characterization of the Human and Rat Islet-Specific Glucose-6-Phosphatase Catalytic Subunit-Related Protein (IGRP) Genes. J Biol Chem 276, 25197–25207 - PubMed
-
- Xin Y, Kim J, Okamoto H, Ni M, Wei Y, Adler C, Murphy AJ, Yancopoulos GD, Lin C, and Gromada J (2016) RNA Sequencing of Single Human Islet Cells Reveals Type 2 Diabetes Genes. Cell Metab 24, 608–615 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

