NnARF17 and NnARF18 from lotus promote root formation and modulate stress tolerance in transgenic Arabidopsis thaliana
- PMID: 38431568
- PMCID: PMC10908128
- DOI: 10.1186/s12870-024-04852-9
NnARF17 and NnARF18 from lotus promote root formation and modulate stress tolerance in transgenic Arabidopsis thaliana
Abstract
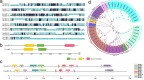
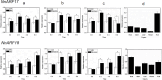
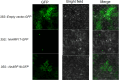
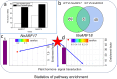
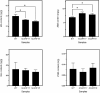
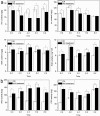
Auxin response factors (ARFs) play a crucial role in regulating gene expression within the auxin signal transduction pathway, particularly during adventitious root (AR) formation. In this investigation, we identified full-length sequences for ARF17 and ARF18, encompassing 1,800 and 2,055 bp, encoding 599 and 684 amino acid residues, respectively. Despite exhibiting low sequence homology, the ARF17- and ARF18-encoded proteins displayed significant structural similarity and shared identical motifs. Phylogenetic analysis revealed close relationships between NnARF17 and VvARF17, as well as NnARF18 and BvARF18. Both ARF17 and ARF18 demonstrated responsiveness to exogenous indole-3-acetic acid (IAA), ethephon, and sucrose, exhibiting organ-specific expression patterns. Beyond their role in promoting root development, these ARFs enhanced stem growth and conferred drought tolerance while mitigating waterlogging stress in transgenic Arabidopsis plants. RNA sequencing data indicated upregulation of 51 and 75 genes in ARF17 and ARF18 transgenic plants, respectively, including five and three genes associated with hormone metabolism and responses. Further analysis of transgenic plants revealed a significant decrease in IAA content, accompanied by a marked increase in abscisic acid content under normal growth conditions. Additionally, lotus seedlings treated with IAA exhibited elevated levels of polyphenol oxidase, IAA oxidase, and peroxidase. The consistent modulation of IAA content in both lotus and transgenic plants highlights the pivotal role of IAA in AR formation in lotus seedlings.
Keywords: Arabidopsis; NnARF17; NnARF18; Adventitious root; lotus.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

