A GREB1-steroid receptor feedforward mechanism governs differential GREB1 action in endometrial function and endometriosis
- PMID: 38431630
- PMCID: PMC10908778
- DOI: 10.1038/s41467-024-46180-4
A GREB1-steroid receptor feedforward mechanism governs differential GREB1 action in endometrial function and endometriosis
Abstract
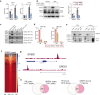
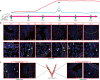
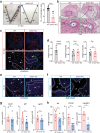
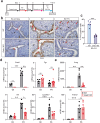
Cellular responses to the steroid hormones, estrogen (E2), and progesterone (P4) are governed by their cognate receptor's transcriptional output. However, the feed-forward mechanisms that shape cell-type-specific transcriptional fulcrums for steroid receptors are unidentified. Herein, we found that a common feed-forward mechanism between GREB1 and steroid receptors regulates the differential effect of GREB1 on steroid hormones in a physiological or pathological context. In physiological (receptive) endometrium, GREB1 controls P4-responses in uterine stroma, affecting endometrial receptivity and decidualization, while not affecting E2-mediated epithelial proliferation. Of mechanism, progesterone-induced GREB1 physically interacts with the progesterone receptor, acting as a cofactor in a positive feedback mechanism to regulate P4-responsive genes. Conversely, in endometrial pathology (endometriosis), E2-induced GREB1 modulates E2-dependent gene expression to promote the growth of endometriotic lesions in mice. This differential action of GREB1 exerted by a common feed-forward mechanism with steroid receptors advances our understanding of mechanisms that underlie cell- and tissue-specific steroid hormone actions.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

