Lopinavir enhances anoikis by remodeling autophagy in a circRNA-dependent manner
- PMID: 38433354
- PMCID: PMC11210930
- DOI: 10.1080/15548627.2024.2325304
Lopinavir enhances anoikis by remodeling autophagy in a circRNA-dependent manner
Abstract
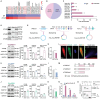
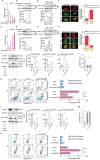
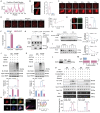
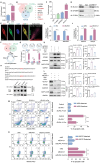
Macroautophagy/autophagy-mediated anoikis resistance is crucial for tumor metastasis. As a key autophagy-related protein, ATG4B has been demonstrated to be a prospective anti-tumor target. However, the existing ATG4B inhibitors are still far from clinical application, especially for tumor metastasis. In this study, we identified a novel circRNA, circSPECC1, that interacted with ATG4B. CircSPECC1 facilitated liquid-liquid phase separation of ATG4B, which boosted the ubiquitination and degradation of ATG4B in gastric cancer (GC) cells. Thus, pharmacological addition of circSPECC1 may serve as an innovative approach to suppress autophagy by targeting ATG4B. Specifically, the circSPECC1 underwent significant m6A modification in GC cells and was subsequently recognized and suppressed by the m6A reader protein ELAVL1/HuR. The activation of the ELAVL1-circSPECC1-ATG4B pathway was demonstrated to mediate anoikis resistance in GC cells. Moreover, we also verified that the above pathway was closely related to metastasis in tissues from GC patients. Furthermore, we determined that the FDA-approved compound lopinavir efficiently enhanced anoikis and prevented metastasis by eliminating repression of ELAVL1 on circSPECC1. In summary, this study provides novel insights into ATG4B-mediated autophagy and introduces a viable clinical inhibitor of autophagy, which may be beneficial for the treatment of GC with metastasis.
Keywords: ATG4B; ELAVL1; circSPECC1; gastric cancer; m6A; metastasis.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures



Comment in
-
CircRNA regulates the liquid-liquid phase separation of ATG4B, a novel strategy to inhibit cancer metastasis?Cell Stress. 2024 May 24;8:56-58. doi: 10.15698/cst2024.05.296. eCollection 2024. Cell Stress. 2024. PMID: 38803355 Free PMC article.
Similar articles
-
CircRNA regulates the liquid-liquid phase separation of ATG4B, a novel strategy to inhibit cancer metastasis?Cell Stress. 2024 May 24;8:56-58. doi: 10.15698/cst2024.05.296. eCollection 2024. Cell Stress. 2024. PMID: 38803355 Free PMC article.
-
Circ-0000953 deficiency exacerbates podocyte injury and autophagy disorder by targeting Mir665-3p-Atg4b in diabetic nephropathy.Autophagy. 2024 May;20(5):1072-1097. doi: 10.1080/15548627.2023.2286128. Epub 2023 Dec 5. Autophagy. 2024. PMID: 38050963 Free PMC article.
-
Targeting the MIR34C-5p-ATG4B-autophagy axis enhances the sensitivity of cervical cancer cells to pirarubicin.Autophagy. 2016 Jul 2;12(7):1105-17. doi: 10.1080/15548627.2016.1173798. Epub 2016 Apr 20. Autophagy. 2016. PMID: 27097054 Free PMC article.
-
On ATG4B as Drug Target for Treatment of Solid Tumours-The Knowns and the Unknowns.Cells. 2019 Dec 24;9(1):53. doi: 10.3390/cells9010053. Cells. 2019. PMID: 31878323 Free PMC article. Review.
-
Targeting Atg4B for cancer therapy: Chemical mediators.Eur J Med Chem. 2021 Jan 1;209:112917. doi: 10.1016/j.ejmech.2020.112917. Epub 2020 Oct 11. Eur J Med Chem. 2021. PMID: 33077263 Review.
Cited by
-
Anoikis resistance in gastric cancer: a comprehensive review.Cell Death Dis. 2025 Jul 15;16(1):528. doi: 10.1038/s41419-025-07860-1. Cell Death Dis. 2025. PMID: 40664635 Free PMC article. Review.
-
Antiretroviral Drugs Impact Autophagy: Opportunities for Drug Repurposing.Front Biosci (Landmark Ed). 2024 Jul 2;29(7):242. doi: 10.31083/j.fbl2907242. Front Biosci (Landmark Ed). 2024. PMID: 39082334 Free PMC article. Review.
-
Identification of Anoikis-Related Genes in Gastric Cancer: Bioinformatics and Experimental Validation.Cancer Med. 2025 Apr;14(8):e70907. doi: 10.1002/cam4.70907. Cancer Med. 2025. PMID: 40263929 Free PMC article.
-
PRSS23-eIF4E-c-Myc axis promotes gastric tumorigenesis and progression.Oncogene. 2025 May;44(17):1167-1181. doi: 10.1038/s41388-025-03294-3. Epub 2025 Feb 7. Oncogene. 2025. PMID: 39920289
-
Biologic activity and treatment resistance to gastrointestinal cancer: the role of circular RNA in autophagy regulation.Front Oncol. 2024 Aug 30;14:1393670. doi: 10.3389/fonc.2024.1393670. eCollection 2024. Front Oncol. 2024. PMID: 39281375 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
