A genetic screen identifies C. elegans eif-3.H and hrpr-1 as pro-apoptotic genes and potential activators of egl-1 expression
- PMID: 38434221
- PMCID: PMC10905296
- DOI: 10.17912/micropub.biology.001126
A genetic screen identifies C. elegans eif-3.H and hrpr-1 as pro-apoptotic genes and potential activators of egl-1 expression
Abstract
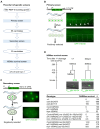
During C. elegans development, 1090 somatic cells are generated of which 131 reproducibly die, many through apoptosis. The C. elegans BH3-only gene egl-1 is the key activator of apoptosis in somatic tissues, and it is predominantly expressed in 'cell death' lineages i.e. lineages in which apoptotic cell death occurs. egl-1 expression is regulated at the transcriptional and post-transcriptional level. For example, we previously showed that the miR-35 and miR-58 families of miRNAs repress egl-1 expression in mothers of 'unwanted' cells by binding to the 3' UTR of egl-1 mRNA, thereby increasing egl-1 mRNA turnover. In a screen for RNA-binding proteins with a role in the post-transcriptional control of egl-1 expression, we identified EIF-3.H (ortholog of human eIF3H) and HRPR-1 (ortholog human hnRNP R/Q) as potential activators of egl-1 expression. In addition, we demonstrate that the knockdown of the eif-3.H or hrpr-1 gene by RNA-mediated interference (RNAi) results in the inappropriate survival of unwanted cells during C. elegans development. Our study provides novel insight into how egl-1 expression is controlled to cause the reproducible pattern of cell death observed during C. elegans development.
Copyright: © 2024 by the authors.
Conflict of interest statement
The authors declare that there are no conflicts of interest present.
Figures
Similar articles
-
miRNAs cooperate in apoptosis regulation during C. elegans development.Genes Dev. 2017 Jan 15;31(2):209-222. doi: 10.1101/gad.288555.116. Epub 2017 Feb 6. Genes Dev. 2017. PMID: 28167500 Free PMC article.
-
PUF-8, a C. elegans ortholog of the RNA-binding proteins PUM1 and PUM2, is required for robustness of the cell death fate.Development. 2023 Oct 1;150(19):dev201167. doi: 10.1242/dev.201167. Epub 2023 Oct 6. Development. 2023. PMID: 37747106 Free PMC article.
-
Timing of the onset of a developmental cell death is controlled by transcriptional induction of the C. elegans ced-3 caspase-encoding gene.Development. 2007 Apr;134(7):1357-68. doi: 10.1242/dev.02818. Epub 2007 Feb 28. Development. 2007. PMID: 17329362
-
egl-1: a key activator of apoptotic cell death in C. elegans.Oncogene. 2008 Dec;27 Suppl 1:S30-40. doi: 10.1038/onc.2009.41. Oncogene. 2008. PMID: 19641505 Review.
-
Keeping killers on a tight leash: transcriptional and post-translational control of the pro-apoptotic activity of BH3-only proteins.Cell Death Differ. 2002 May;9(5):505-12. doi: 10.1038/sj.cdd.4400998. Cell Death Differ. 2002. PMID: 11973609 Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
