Photo-addressable microwell devices for rapid functional screening and isolation of pathogen inhibitors from bacterial strain libraries
- PMID: 38434239
- PMCID: PMC10907074
- DOI: 10.1063/5.0188270
Photo-addressable microwell devices for rapid functional screening and isolation of pathogen inhibitors from bacterial strain libraries
Abstract
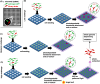
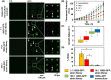
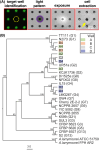
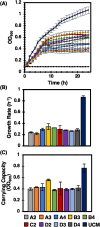
Discovery of new strains of bacteria that inhibit pathogen growth can facilitate improvements in biocontrol and probiotic strategies. Traditional, plate-based co-culture approaches that probe microbial interactions can impede this discovery as these methods are inherently low-throughput, labor-intensive, and qualitative. We report a second-generation, photo-addressable microwell device, developed to iteratively screen interactions between candidate biocontrol agents existing in bacterial strain libraries and pathogens under increasing pathogen pressure. Microwells (0.6 pl volume) provide unique co-culture sites between library strains and pathogens at controlled cellular ratios. During sequential screening iterations, library strains are challenged against increasing numbers of pathogens to quantitatively identify microwells containing strains inhibiting the highest numbers of pathogens. Ring-patterned 365 nm light is then used to ablate a photodegradable hydrogel membrane and sequentially release inhibitory strains from the device for recovery. Pathogen inhibition with each recovered strain is validated, followed by whole genome sequencing. To demonstrate the rapid nature of this approach, the device was used to screen a 293-membered biovar 1 agrobacterial strain library for strains inhibitory to the plant pathogen Agrobacterium tumefaciens sp. 15955. One iterative screen revealed nine new inhibitory strains. For comparison, plate-based methods did not uncover any inhibitory strains from the library (n = 30 plates). The novel pathogen-challenge screening mode developed here enables rapid selection and recovery of strains that effectively suppress pathogen growth from bacterial strain libraries, expanding this microwell technology platform toward rapid, cost-effective, and scalable screening for probiotics, biocontrol agents, and inhibitory molecules that can protect against known or emerging pathogens.
© 2024 Author(s).
Conflict of interest statement
R.R.H. and T.G.P. have a patent on this technology (U.S. Patent No. 11,235,330).57 The other authors have no conflicts to disclose.
Figures

References
-
- Chaloner T. M., Gurr S. J., and Bebber D. P., Nature Climate Change 11(8), 710–715 (2021). 10.1038/s41558-021-01104-8 - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
