SH2B1 Tunes Hippocampal ERK Signaling to Influence Fluid Intelligence in Humans and Mice
- PMID: 38434247
- PMCID: PMC10907025
- DOI: 10.34133/research.0269
SH2B1 Tunes Hippocampal ERK Signaling to Influence Fluid Intelligence in Humans and Mice
Abstract
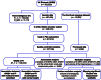
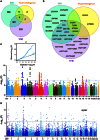
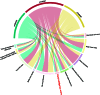
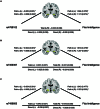
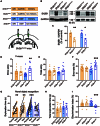
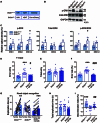
Fluid intelligence is a cognitive domain that encompasses general reasoning, pattern recognition, and problem-solving abilities independent of task-specific experience. Understanding its genetic and neural underpinnings is critical yet challenging for predicting human development, lifelong health, and well-being. One approach to address this challenge is to map the network of correlations between intelligence and other constructs. In the current study, we performed a genome-wide association study using fluid intelligence quotient scores from the UK Biobank to explore the genetic architecture of the associations between obesity risk and fluid intelligence. Our results revealed novel common genetic loci (SH2B1, TUFM, ATP2A1, and FOXO3) underlying the association between fluid intelligence and body metabolism. Surprisingly, we demonstrated that SH2B1 variation influenced fluid intelligence independently of its effects on metabolism but partially mediated its association with bilateral hippocampal volume. Consistently, selective genetic ablation of Sh2b1 in the mouse hippocampus, particularly in inhibitory neurons, but not in excitatory neurons, significantly impaired working memory, short-term novel object recognition memory, and behavioral flexibility, but not spatial learning and memory, mirroring the human intellectual performance. Single-cell genetic profiling of Sh2B1-regulated molecular pathways revealed that Sh2b1 deletion resulted in aberrantly enhanced extracellular signal-regulated kinase (ERK) signaling, whereas pharmacological inhibition of ERK signaling reversed the associated behavioral impairment. Our cross-species study thus provides unprecedented insight into the role of SH2B1 in fluid intelligence and has implications for understanding the genetic and neural underpinnings of lifelong mental health and well-being.
Copyright © 2023 Xiujuan Du et al.
Conflict of interest statement
Competing interests: The authors declare that they have no competing interests.
Figures


Similar articles
-
Nociceptin receptor impairs recognition memory via interaction with NMDA receptor-dependent mitogen-activated protein kinase/extracellular signal-regulated kinase signaling in the hippocampus.J Neurosci. 2008 Feb 27;28(9):2190-8. doi: 10.1523/JNEUROSCI.3711-07.2008. J Neurosci. 2008. PMID: 18305252 Free PMC article.
-
17β-Estradiol and Agonism of G-protein-Coupled Estrogen Receptor Enhance Hippocampal Memory via Different Cell-Signaling Mechanisms.J Neurosci. 2016 Mar 16;36(11):3309-21. doi: 10.1523/JNEUROSCI.0257-15.2016. J Neurosci. 2016. PMID: 26985039 Free PMC article.
-
The SH2B1 obesity locus and abnormal glucose homeostasis: lack of evidence for association from a meta-analysis in individuals of European ancestry.Nutr Metab Cardiovasc Dis. 2013 Nov;23(11):1043-9. doi: 10.1016/j.numecd.2013.05.001. Epub 2013 Oct 5. Nutr Metab Cardiovasc Dis. 2013. PMID: 24103803
-
SH2B1 regulation of energy balance, body weight, and glucose metabolism.World J Diabetes. 2014 Aug 15;5(4):511-26. doi: 10.4239/wjd.v5.i4.511. World J Diabetes. 2014. PMID: 25126397 Free PMC article. Review.
-
New perspective on SH2B1: An accelerator of cancer progression.Biomed Pharmacother. 2020 Jan;121:109651. doi: 10.1016/j.biopha.2019.109651. Epub 2019 Nov 15. Biomed Pharmacother. 2020. PMID: 31739166 Review.
Cited by
-
Genome-wide Pleiotropy Analysis Reveals Shared Genetic Associations between Type 2 Diabetes Mellitus and Subcortical Brain Volumes.Research (Wash D C). 2025 May 6;8:0688. doi: 10.34133/research.0688. eCollection 2025. Research (Wash D C). 2025. PMID: 40330659 Free PMC article.
-
TUFM in health and disease: exploring its multifaceted roles.Front Immunol. 2024 May 29;15:1424385. doi: 10.3389/fimmu.2024.1424385. eCollection 2024. Front Immunol. 2024. PMID: 38868764 Free PMC article. Review.
References
-
- Deary IJ, Penke L, Johnson W. The neuroscience of human intelligence differences. Nat Rev Neurosci. 2010;11(3):201–211. - PubMed
-
- Bartels M, Rietveld MJ, Van Baal, Boomsma DI. Genetic and environmental influences on the development of intelligence. Behav Genet. 2002;32(4):237–249. - PubMed
-
- Wisdom NM, Callahan JL, Hawkins KA. The effects of apolipoprotein E on non-impaired cognitive functioning: A meta-analysis. Neurobiol Aging. 2011;32(1):63–74. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

