DrosOMA: the Drosophila Orthologous Matrix browser
- PMID: 38434623
- PMCID: PMC10905159
- DOI: 10.12688/f1000research.135250.2
DrosOMA: the Drosophila Orthologous Matrix browser
Abstract
Background: Comparative genomic analyses to delineate gene evolutionary histories inform the understanding of organismal biology by characterising gene and gene family origins, trajectories, and dynamics, as well as enabling the tracing of speciation, duplication, and loss events, and facilitating the transfer of gene functional information across species. Genomic data are available for an increasing number of species from the genus Drosophila, however, a dedicated resource exploiting these data to provide the research community with browsable results from genus-wide orthology delineation has been lacking.
Methods: Using the OMA Orthologous Matrix orthology inference approach and browser deployment framework, we catalogued orthologues across a selected set of Drosophila species with high-quality annotated genomes. We developed and deployed a dedicated instance of the OMA browser to facilitate intuitive exploration, visualisation, and downloading of the genus-wide orthology delineation results.
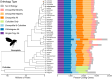
Results: DrosOMA - the Drosophila Orthologous Matrix browser, accessible from https://drosoma.dcsr.unil.ch/ - presents the results of orthology delineation for 36 drosophilids from across the genus and four outgroup dipterans. It enables querying and browsing of the orthology data through a feature-rich web interface, with gene-view, orthologous group-view, and genome-view pages, including comprehensive gene name and identifier cross-references together with available functional annotations and protein domain architectures, as well as tools to visualise local and global synteny conservation.
Conclusions: The DrosOMA browser demonstrates the deployability of the OMA browser framework for building user-friendly orthology databases with dense sampling of a selected taxonomic group. It provides the Drosophila research community with a tailored resource of browsable results from genus-wide orthology delineation.
Keywords: Drosophila; comparative genomics; database; gene families; orthologous groups; orthologues; orthology; synteny.
Copyright: © 2024 Thiébaut A et al.
Conflict of interest statement
No competing interests were disclosed.
Figures


References
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

