Predicting pharmacodynamic effects through early drug discovery with artificial intelligence-physiologically based pharmacokinetic (AI-PBPK) modelling
- PMID: 38434709
- PMCID: PMC10904617
- DOI: 10.3389/fphar.2024.1330855
Predicting pharmacodynamic effects through early drug discovery with artificial intelligence-physiologically based pharmacokinetic (AI-PBPK) modelling
Abstract
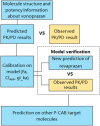
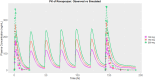
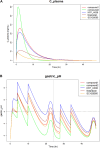
A mechanism-based pharmacokinetic/pharmacodynamic (PK/PD) model links the concentration-time profile of a drug with its therapeutic effects based on the underlying biological or physiological processes. Clinical endpoints play a pivotal role in drug development. Despite the substantial time and effort invested in screening drugs for favourable pharmacokinetic (PK) properties, they may not consistently yield optimal clinical outcomes. Furthermore, in the virtual compound screening phase, researchers cannot observe clinical outcomes in humans directly. These uncertainties prolong the process of drug development. As incorporation of Artificial Intelligence (AI) into the physiologically based pharmacokinetic/pharmacodynamic (PBPK) model can assist in forecasting pharmacodynamic (PD) effects within the human body, we introduce a methodology for utilizing the AI-PBPK platform to predict the PK and PD outcomes of target compounds in the early drug discovery stage. In this integrated platform, machine learning is used to predict the parameters for the model, and the mechanism-based PD model is used to predict the PD outcome through the PK results. This platform enables researchers to align the PK profile of a drug with desired PD effects at the early drug discovery stage. Case studies are presented to assess and compare five potassium-competitive acid blocker (P-CAB) compounds, after calibration and verification using vonoprazan and revaprazan.
Keywords: P-CABs; PBPK modeling; PD modeling; artificial intelligence (AI); early drug discovery; machine learning (ML).
Copyright © 2024 Wu, Li, Zhou, Zhao, Su, Cheng, Wu, Huang, Jin, Li, Zhang, Liu and Liu.
Conflict of interest statement
KW, XL, ZZ, YZ, ZC, XW, ZH, and JaL were employees of the Yinghan Pharmaceutical Technology (Shanghai) at the time of study conduct. MS was the employee of the Jiangsu Carephar Pharmaceutical Co., Ltd. at the time of study conduct. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Prediction of pharmacokinetic/pharmacodynamic properties of aldosterone synthase inhibitors at drug discovery stage using an artificial intelligence-physiologically based pharmacokinetic model.Front Pharmacol. 2025 Apr 28;16:1578117. doi: 10.3389/fphar.2025.1578117. eCollection 2025. Front Pharmacol. 2025. PMID: 40356995 Free PMC article.
-
Machine learning and artificial intelligence in physiologically based pharmacokinetic modeling.Toxicol Sci. 2023 Jan 31;191(1):1-14. doi: 10.1093/toxsci/kfac101. Toxicol Sci. 2023. PMID: 36156156 Free PMC article.
-
An Integrated AI-PBPK Platform for Predicting Drug In Vivo Fate and Tissue Distribution in Human and Inter-Species Extrapolation.Clin Pharmacol Ther. 2025 May 26. doi: 10.1002/cpt.3732. Online ahead of print. Clin Pharmacol Ther. 2025. PMID: 40418625
-
Physiologically-based PK/PD modelling of therapeutic macromolecules.Pharm Res. 2009 Dec;26(12):2543-50. doi: 10.1007/s11095-009-9990-3. Epub 2009 Oct 22. Pharm Res. 2009. PMID: 19847627 Review.
-
The Role of Pharmacometrics in Advancing the Therapies for Autoimmune Diseases.Pharmaceutics. 2024 Dec 5;16(12):1559. doi: 10.3390/pharmaceutics16121559. Pharmaceutics. 2024. PMID: 39771538 Free PMC article. Review.
Cited by
-
The Potential of Artificial Intelligence in Pharmaceutical Innovation: From Drug Discovery to Clinical Trials.Pharmaceuticals (Basel). 2025 May 25;18(6):788. doi: 10.3390/ph18060788. Pharmaceuticals (Basel). 2025. PMID: 40573185 Free PMC article. Review.
-
Computational identification of novel natural inhibitors against triple mutant DNA gyrase A in fluoroquinolone-resistant Salmonella Typhimurium.Biochem Biophys Rep. 2025 Jan 8;41:101901. doi: 10.1016/j.bbrep.2024.101901. eCollection 2025 Mar. Biochem Biophys Rep. 2025. PMID: 39867681 Free PMC article.
-
Prediction of pharmacokinetic/pharmacodynamic properties of aldosterone synthase inhibitors at drug discovery stage using an artificial intelligence-physiologically based pharmacokinetic model.Front Pharmacol. 2025 Apr 28;16:1578117. doi: 10.3389/fphar.2025.1578117. eCollection 2025. Front Pharmacol. 2025. PMID: 40356995 Free PMC article.
-
Risks of anti-Helicobacter therapy and long-term therapy with antisecretory drugs.World J Gastroenterol. 2025 Jan 28;31(4):101933. doi: 10.3748/wjg.v31.i4.101933. World J Gastroenterol. 2025. PMID: 39877710 Free PMC article.
-
Predictive Modeling of Pharmacokinetic Drug-Drug and Herb-Drug Interactions in Oncology: Insights From PBPK Studies.Int J Toxicol. 2025 Sep-Oct;44(5):424-440. doi: 10.1177/10915818251345116. Epub 2025 Jun 11. Int J Toxicol. 2025. PMID: 40499171 Free PMC article. Review.
References
-
- Gilmer J., Riley P. F., Oriol V., Dahl G. E. (2017). Neural message passing for quantum chemistry. https://arxiv.org/abs/1704.01212.
LinkOut - more resources
Full Text Sources
Research Materials

