The genome sequence of the Wasp Spider, Argiope bruennichi (Scopoli, 1772)
- PMID: 38434739
- PMCID: PMC10904950
- DOI: 10.12688/wellcomeopenres.20339.1
The genome sequence of the Wasp Spider, Argiope bruennichi (Scopoli, 1772)
Abstract
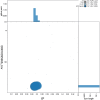
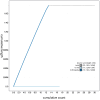
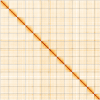
We present a genome assembly from an individual female Argiope bruennichi (the Wasp Spider; Arthropoda; Arachnida; Araneae; Araneidae). The genome sequence is 1,778.4 megabases in span. Most of the assembly is scaffolded into 13 chromosomal pseudomolecules, including the X 1 and X 2 sex chromosomes. The mitochondrial genome has also been assembled and is 14.06 kilobases in length.
Keywords: Araneae; Argiope bruennichi; Wasp Spider; chromosomal; genome sequence.
Copyright: © 2023 Crowley LM et al.
Conflict of interest statement
No competing interests were disclosed.
Figures


Similar articles
-
The genome sequence of an orbweaving spider, Gibbaranea gibbosa (Walckenaer, 1802).Wellcome Open Res. 2025 Feb 24;10:97. doi: 10.12688/wellcomeopenres.23751.1. eCollection 2025. Wellcome Open Res. 2025. PMID: 40162187 Free PMC article.
-
The genome sequence of the silver stretch spider, Tetragnatha montana (Simon, 1874) (Araneae: Tetragnathidae).Wellcome Open Res. 2024 May 22;9:288. doi: 10.12688/wellcomeopenres.21782.1. eCollection 2024. Wellcome Open Res. 2024. PMID: 40012987 Free PMC article.
-
The genome sequence of the Black Lace-weaver spider, Amaurobius ferox (Walckenaer, 1830).Wellcome Open Res. 2024 Mar 1;9:105. doi: 10.12688/wellcomeopenres.21080.1. eCollection 2024. Wellcome Open Res. 2024. PMID: 39015616 Free PMC article.
-
The genome sequence of the spider, Parasteatoda lunata (Clerck, 1757).Wellcome Open Res. 2023 Jun 22;8:271. doi: 10.12688/wellcomeopenres.19585.1. eCollection 2023. Wellcome Open Res. 2023. PMID: 37766855 Free PMC article.
-
The genome sequence of the Common Sheetweb Spider Linyphia triangularis (Clerck, 1757).Wellcome Open Res. 2025 Feb 24;10:92. doi: 10.12688/wellcomeopenres.23754.1. eCollection 2025. Wellcome Open Res. 2025. PMID: 40084296 Free PMC article.
References
-
- Bee L, Oxford G, Smith H: Britain’s Spiders.2nd ed. Princeton University Press,2020. 10.1515/9780691211800 - DOI
-
- British Arachnological Society: Spider and Harvestman Recording Scheme website: Summary for Argiope bruennichi. (Araneae).2023; [Accessed 12 October 2023]. Reference Source
Grants and funding
LinkOut - more resources
Full Text Sources

