Biophysical Analysis of Potential Inhibitors of SARS-CoV-2 Cell Recognition and Their Effect on Viral Dynamics in Different Cell Types: A Computational Prediction from In Vitro Experimental Data
- PMID: 38434903
- PMCID: PMC10905729
- DOI: 10.1021/acsomega.3c06968
Biophysical Analysis of Potential Inhibitors of SARS-CoV-2 Cell Recognition and Their Effect on Viral Dynamics in Different Cell Types: A Computational Prediction from In Vitro Experimental Data
Abstract
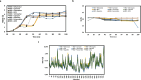
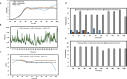
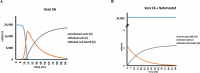
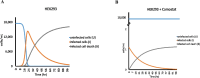
Recent reports have suggested that the susceptibility of cells to SARS-CoV-2 infection can be influenced by various proteins that potentially act as receptors for the virus. To investigate this further, we conducted simulations of viral dynamics using different cellular systems (Vero E6, HeLa, HEK293, and CaLu3) in the presence and absence of drugs (anthelmintic, ARBs, anticoagulant, serine protease inhibitor, antimalarials, and NSAID) that have been shown to impact cellular recognition by the spike protein based on experimental data. Our simulations revealed that the susceptibility of the simulated cell systems to SARS-CoV-2 infection was similar across all tested systems. Notably, CaLu3 cells exhibited the highest susceptibility to SARS-CoV-2 infection, potentially due to the presence of receptors other than ACE2, which may account for a significant portion of the observed susceptibility. Throughout the study, all tested compounds showed thermodynamically favorable and stable binding to the spike protein. Among the tested compounds, the anticoagulant nafamostat demonstrated the most favorable characteristics in terms of thermodynamics, kinetics, theoretical antiviral activity, and potential safety (toxicity) in relation to SARS-CoV-2 spike protein-mediated infections in the tested cell lines. This study provides mathematical and bioinformatic models that can aid in the identification of optimal cell lines for compound evaluation and detection, particularly in studies focused on repurposed drugs and their mechanisms of action. It is important to note that these observations should be experimentally validated, and this research is expected to inspire future quantitative experiments.
© 2024 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

Similar articles
-
Comparative study of SARS-CoV-2 infection in different cell types: Biophysical-computational approach to the role of potential receptors.Comput Biol Med. 2022 Mar;142:105245. doi: 10.1016/j.compbiomed.2022.105245. Epub 2022 Jan 20. Comput Biol Med. 2022. PMID: 35077937 Free PMC article.
-
Distinctive Roles of Furin and TMPRSS2 in SARS-CoV-2 Infectivity.J Virol. 2022 Apr 27;96(8):e0012822. doi: 10.1128/jvi.00128-22. Epub 2022 Mar 28. J Virol. 2022. PMID: 35343766 Free PMC article.
-
The Anticoagulant Nafamostat Potently Inhibits SARS-CoV-2 S Protein-Mediated Fusion in a Cell Fusion Assay System and Viral Infection In Vitro in a Cell-Type-Dependent Manner.Viruses. 2020 Jun 10;12(6):629. doi: 10.3390/v12060629. Viruses. 2020. PMID: 32532094 Free PMC article.
-
Potential therapeutic approaches for the early entry of SARS-CoV-2 by interrupting the interaction between the spike protein on SARS-CoV-2 and angiotensin-converting enzyme 2 (ACE2).Biochem Pharmacol. 2021 Oct;192:114724. doi: 10.1016/j.bcp.2021.114724. Epub 2021 Aug 8. Biochem Pharmacol. 2021. PMID: 34371003 Free PMC article. Review.
-
Therapeutic potential of green tea catechin, (-)-epigallocatechin-3-O-gallate (EGCG) in SARS-CoV-2 infection: Major interactions with host/virus proteases.Phytomed Plus. 2023 Feb;3(1):100402. doi: 10.1016/j.phyplu.2022.100402. Epub 2022 Dec 30. Phytomed Plus. 2023. PMID: 36597465 Free PMC article. Review.
Cited by
-
Biological Implications of the Intrinsic Deformability of Human Acetylcholinesterase Induced by Diverse Compounds: A Computational Study.Biology (Basel). 2024 Dec 19;13(12):1065. doi: 10.3390/biology13121065. Biology (Basel). 2024. PMID: 39765732 Free PMC article.
References
-
- Kyrou I.; Randeva H. S.; Spandidos D. A.; Karteris E. Not only ACE2—the quest for additional host cell mediators of SARS-CoV-2 infection: Neuropilin-1 (NRP1) as a novel SARS-CoV-2 host cell entry mediator implicated in COVID-19. Signal transduction and targeted therapy 2021, 6 (1), 1–3. 10.1038/s41392-020-00460-9. - DOI - PMC - PubMed
-
- Dobrindt K.; Hoagland D. A.; Seah C.; Kassim B.; O'Shea C. P.; Murphy A.; Iskhakova M.; Fernando M. B.; Powell S. K.; Deans P. J. M.; Javidfar B.; Peter C.; Møller R.; Uhl S. A.; Garcia M. F.; Kimura M.; Iwasawa K.; Crary J. F.; Kotton D. N.; Takebe T.; Huckins L. M.; tenOever B. R.; Akbarian S.; Brennand K. J. Common genetic variation in humans impacts in vitro susceptibility to SARS-CoV-2 infection. Stem Cell Rep. 2021, 16 (3), 505–518. 10.1016/j.stemcr.2021.02.010. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
