Cellular Repair of Synthetic Analogs of Oxidative DNA Damage Reveals a Key Structure-Activity Relationship of the Cancer-Associated MUTYH DNA Repair Glycosylase
- PMID: 38435525
- PMCID: PMC10906249
- DOI: 10.1021/acscentsci.3c00784
Cellular Repair of Synthetic Analogs of Oxidative DNA Damage Reveals a Key Structure-Activity Relationship of the Cancer-Associated MUTYH DNA Repair Glycosylase
Abstract
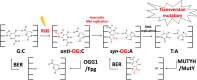
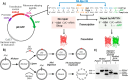
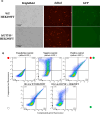
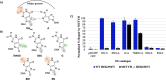
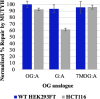
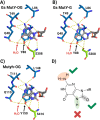
The base excision repair glycosylase MUTYH prevents mutations associated with the oxidatively damaged base, 8-oxo-7,8-dihydroguanine (OG), by removing undamaged misincorporated adenines from OG:A mispairs. Defects in OG:A repair in individuals with inherited MUTYH variants are correlated with the colorectal cancer predisposition syndrome known as MUTYH-associated polyposis (MAP). Herein, we reveal key structural features of OG required for efficient repair by human MUTYH using structure-activity relationships (SAR). We developed a GFP-based plasmid reporter assay to define SAR with synthetically generated OG analogs in human cell lines. Cellular repair results were compared with kinetic parameters measured by adenine glycosylase assays in vitro. Our results show substrates lacking the 2-amino group of OG, such as 8OI:A (8OI = 8-oxoinosine), are not repaired in cells, despite being excellent substrates in in vitro adenine glycosylase assays, new evidence that the search and detection steps are critical factors in cellular MUTYH repair functionality. Surprisingly, modification of the O8/N7H of OG, which is the distinguishing feature of OG relative to G, was tolerated in both MUTYH-mediated cellular repair and in vitro adenine glycosylase activity. The lack of sensitivity to alterations at the O8/N7H in the SAR of MUTYH substrates is distinct from previous work with bacterial MutY, indicating that the human enzyme is much less stringent in its lesion verification. Our results imply that the human protein relies almost exclusively on detection of the unique major groove position of the 2-amino group of OG within OGsyn:Aanti mispairs to select contextually incorrect adenines for excision and thereby thwart mutagenesis. These results predict that MUTYH variants that exhibit deficiencies in OG:A detection will be severely compromised in a cellular setting. Moreover, the reliance of MUTYH on the interaction with the OG 2-amino group suggests that disrupting this interaction with small molecules may provide a strategy to develop potent and selective MUTYH inhibitors.
© 2024 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
Similar articles
-
FSHing for DNA Damage: Key Features of MutY Detection of 8-Oxoguanine:Adenine Mismatches.Acc Chem Res. 2024 Apr 2;57(7):1019-1031. doi: 10.1021/acs.accounts.3c00759. Epub 2024 Mar 12. Acc Chem Res. 2024. PMID: 38471078 Free PMC article. Review.
-
Detection of OG:A Lesion Mispairs by MutY Relies on a Single His Residue and the 2-Amino Group of 8-Oxoguanine.J Am Chem Soc. 2020 Aug 5;142(31):13283-13287. doi: 10.1021/jacs.0c04284. Epub 2020 Jul 23. J Am Chem Soc. 2020. PMID: 32664726 Free PMC article.
-
Cellular Assays for Studying the Fe-S Cluster Containing Base Excision Repair Glycosylase MUTYH and Homologs.Methods Enzymol. 2018;599:69-99. doi: 10.1016/bs.mie.2017.12.006. Epub 2018 Jan 10. Methods Enzymol. 2018. PMID: 29746250 Free PMC article.
-
Unique Hydrogen Bonding of Adenine with the Oxidatively Damaged Base 8-Oxoguanine Enables Specific Recognition and Repair by DNA Glycosylase MutY.J Am Chem Soc. 2020 Dec 2;142(48):20340-20350. doi: 10.1021/jacs.0c06767. Epub 2020 Nov 17. J Am Chem Soc. 2020. PMID: 33202125 Free PMC article.
-
When you're strange: Unusual features of the MUTYH glycosylase and implications in cancer.DNA Repair (Amst). 2019 Aug;80:16-25. doi: 10.1016/j.dnarep.2019.05.005. Epub 2019 Jun 8. DNA Repair (Amst). 2019. PMID: 31203172 Free PMC article. Review.
Cited by
-
Structure of human MUTYH and functional profiling of cancer-associated variants reveal an allosteric network between its [4Fe-4S] cluster cofactor and active site required for DNA repair.Nat Commun. 2025 Apr 16;16(1):3596. doi: 10.1038/s41467-025-58361-w. Nat Commun. 2025. PMID: 40234396 Free PMC article.
-
Precision genome editing and in-cell measurements of oxidative DNA damage repair enable functional and mechanistic characterization of cancer-associated MUTYH variants.Nucleic Acids Res. 2025 Mar 20;53(6):gkaf037. doi: 10.1093/nar/gkaf037. Nucleic Acids Res. 2025. PMID: 40156857 Free PMC article.
-
FSHing for DNA Damage: Key Features of MutY Detection of 8-Oxoguanine:Adenine Mismatches.Acc Chem Res. 2024 Apr 2;57(7):1019-1031. doi: 10.1021/acs.accounts.3c00759. Epub 2024 Mar 12. Acc Chem Res. 2024. PMID: 38471078 Free PMC article. Review.
-
Crystal structure of MutYX: A novel clusterless adenine DNA glycosylase with a distinct C-terminal domain and 8-Oxoguanine recognition sphere.bioRxiv [Preprint]. 2025 Jan 3:2025.01.03.631205. doi: 10.1101/2025.01.03.631205. bioRxiv. 2025. PMID: 39803464 Free PMC article. Preprint.
-
Saturation mapping of MUTYH variant effects using DNA repair reporters.bioRxiv [Preprint]. 2025 Mar 6:2025.03.01.640912. doi: 10.1101/2025.03.01.640912. bioRxiv. 2025. Update in: Am J Hum Genet. 2025 Jul 24:S0002-9297(25)00275-7. doi: 10.1016/j.ajhg.2025.07.005. PMID: 40093110 Free PMC article. Updated. Preprint.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
