Vegetative cell wall protein OsGP1 regulates cell wall mediated soda saline-alkali stress in rice
- PMID: 38436004
- PMCID: PMC10908258
- DOI: 10.7717/peerj.16790
Vegetative cell wall protein OsGP1 regulates cell wall mediated soda saline-alkali stress in rice
Abstract
Plant growth and development are inhibited by the high levels of ions and pH due to soda saline-alkali soil, and the cell wall serves as a crucial barrier against external stresses in plant cells. Proteins in the cell wall play important roles in plant cell growth, morphogenesis, pathogen infection and environmental response. In the current study, the full-length coding sequence of the vegetative cell wall protein gene OsGP1 was characterized from Lj11 (Oryza sativa longjing11), it contained 660 bp nucleotides encoding 219 amino acids. Protein-protein interaction network analysis revealed possible interaction between CESA1, TUBB8, and OsJ_01535 proteins, which are related to plant growth and cell wall synthesis. OsGP1 was found to be localized in the cell membrane and cell wall. Furthermore, overexpression of OsGP1 leads to increase in plant height and fresh weight, showing enhanced resistance to saline-alkali stress. The ROS (reactive oxygen species) scavengers were regulated by OsGP1 protein, peroxidase and superoxide dismutase activities were significantly higher, while malondialdehyde was lower in the overexpression line under stress. These results suggest that OsGP1 improves saline-alkali stress tolerance of rice possibly through cell wall-mediated intracellular environmental homeostasis.
Keywords: Abiotic stress; Gene cloning; Genetic transformation; OsGP1; Rice.
© 2024 Zhu et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
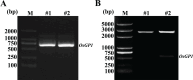





Similar articles
-
OsSAP6 Positively Regulates Soda Saline-Alkaline Stress Tolerance in Rice.Rice (N Y). 2022 Dec 27;15(1):69. doi: 10.1186/s12284-022-00616-x. Rice (N Y). 2022. PMID: 36574073 Free PMC article.
-
Comparative Analysis of Physiological, Hormonal and Transcriptomic Responses Reveal Mechanisms of Saline-Alkali Tolerance in Autotetraploid Rice (Oryza sativa L.).Int J Mol Sci. 2022 Dec 18;23(24):16146. doi: 10.3390/ijms232416146. Int J Mol Sci. 2022. PMID: 36555786 Free PMC article.
-
Alfalfa MsCBL4 enhances calcium metabolism but not sodium transport in transgenic tobacco under salt and saline-alkali stress.Plant Cell Rep. 2020 Aug;39(8):997-1011. doi: 10.1007/s00299-020-02543-x. Epub 2020 Apr 24. Plant Cell Rep. 2020. PMID: 32333150
-
Genome-wide association and selection studies reveal genomic insight into saline-alkali tolerance in rice.Plant J. 2025 Mar;121(6):e70056. doi: 10.1111/tpj.70056. Plant J. 2025. PMID: 40084738
-
Dynamics of cell wall structure and related genomic resources for drought tolerance in rice.Plant Cell Rep. 2021 Mar;40(3):437-459. doi: 10.1007/s00299-020-02649-2. Epub 2021 Jan 2. Plant Cell Rep. 2021. PMID: 33389046 Review.
References
-
- Calderan-Rodrigues MJ, Guimarães Fonseca J, de Moraes FE, Vaz Setem L, Carmanhanis Begossi A, Labate CA. Plant cell wall proteomics: a focus on monocot species, Brachypodium distachyon, Saccharum spp. and Oryza sativa. International Journal of Molecular Sciences. 2019;20(8):1975. doi: 10.3390/ijms20081975. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

