GTPase activity of porcine Mx1 plays a dominant role in inhibiting the N-Nsp9 interaction and thus inhibiting PRRSV replication
- PMID: 38436247
- PMCID: PMC11019876
- DOI: 10.1128/jvi.01844-23
GTPase activity of porcine Mx1 plays a dominant role in inhibiting the N-Nsp9 interaction and thus inhibiting PRRSV replication
Abstract
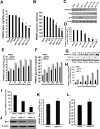
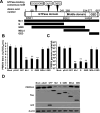
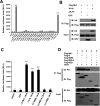
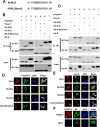
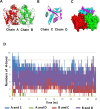
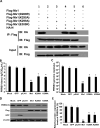
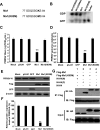
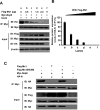
Porcine Mx1 is a type of interferon-induced GTPase that inhibits the replication of certain RNA viruses. However, the antiviral effects and the underlying mechanism of porcine Mx1 for porcine reproductive and respiratory syndrome virus (PRRSV) remain unknown. In this study, we demonstrated that porcine Mx1 could significantly inhibit PRRSV replication in MARC-145 cells. By Mx1 segment analysis, it was indicated that the GTPase domain (68-341aa) was the functional area to inhibit PRRSV replication and that Mx1 interacted with the PRRSV-N protein through the GTPase domain (68-341aa) in the cytoplasm. Amino acid residues K295 and K299 in the G domain of Mx1 were the key sites for Mx1-N interaction while mutant proteins Mx1(K295A) and Mx1(K299A) still partially inhibited PRRSV replication. Furthermore, we found that the GTPase activity of Mx1 was dominant for Mx1 to inhibit PRRSV replication but was not essential for Mx1-N interaction. Finally, mechanistic studies demonstrated that the GTPase activity of Mx1 played a dominant role in inhibiting the N-Nsp9 interaction and that the interaction between Mx1 and N partially inhibited the N-Nsp9 interaction. We propose that the complete anti-PRRSV mechanism of porcine Mx1 contains a two-step process: Mx1 binds to the PRRSV-N protein and subsequently disrupts the N-Nsp9 interaction by a process requiring the GTPase activity of Mx1. Taken together, the results of our experiments describe for the first time a novel mechanism by which porcine Mx1 evolves to inhibit PRRSV replication.
Importance: Mx1 protein is a key mediator of the interferon-induced antiviral response against a wide range of viruses. How porcine Mx1 affects the replication of porcine reproductive and respiratory syndrome virus (PRRSV) and its biological function has not been studied. Here, we show that Mx1 protein inhibits PRRSV replication by interfering with N-Nsp9 interaction. Furthermore, the GTPase activity of porcine Mx1 plays a dominant role and the Mx1-N interaction plays an assistant role in this interference process. This study uncovers a novel mechanism evolved by porcine Mx1 to exert anti-PRRSV activities.
Keywords: Mx1; antiviral agents; interferons; porcine reproductive and respiratory syndrome virus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
ZAP, a CCCH-Type Zinc Finger Protein, Inhibits Porcine Reproductive and Respiratory Syndrome Virus Replication and Interacts with Viral Nsp9.J Virol. 2019 May 1;93(10):e00001-19. doi: 10.1128/JVI.00001-19. Print 2019 May 15. J Virol. 2019. PMID: 30867303 Free PMC article.
-
The interaction of nonstructural protein 9 with retinoblastoma protein benefits the replication of genotype 2 porcine reproductive and respiratory syndrome virus in vitro.Virology. 2014 Sep;464-465:432-440. doi: 10.1016/j.virol.2014.07.036. Epub 2014 Aug 21. Virology. 2014. PMID: 25146601 Free PMC article.
-
An intracellularly expressed Nsp9-specific nanobody in MARC-145 cells inhibits porcine reproductive and respiratory syndrome virus replication.Vet Microbiol. 2015 Dec 31;181(3-4):252-60. doi: 10.1016/j.vetmic.2015.10.021. Epub 2015 Oct 26. Vet Microbiol. 2015. PMID: 26525739
-
Interleukin-2 enhancer binding factor 2 interacts with the nsp9 or nsp2 of porcine reproductive and respiratory syndrome virus and exerts negatively regulatory effect on the viral replication.Virol J. 2017 Jul 11;14(1):125. doi: 10.1186/s12985-017-0794-5. Virol J. 2017. PMID: 28693575 Free PMC article.
-
A Nanobody Targeting Viral Nonstructural Protein 9 Inhibits Porcine Reproductive and Respiratory Syndrome Virus Replication.J Virol. 2019 Feb 5;93(4):e01888-18. doi: 10.1128/JVI.01888-18. Print 2019 Feb 15. J Virol. 2019. PMID: 30463975 Free PMC article.
Cited by
-
Swine NONO promotes IRF3-mediated antiviral immune response by Detecting PRRSV N protein.PLoS Pathog. 2024 Oct 16;20(10):e1012622. doi: 10.1371/journal.ppat.1012622. eCollection 2024 Oct. PLoS Pathog. 2024. PMID: 39413144 Free PMC article.
-
LGALS3BP/90K suppresses porcine reproductive and respiratory syndrome virus replication by enhancing GP3 degradation and stimulating innate immunity.Vet Res. 2025 Jun 20;56(1):121. doi: 10.1186/s13567-025-01556-2. Vet Res. 2025. PMID: 40542445 Free PMC article.
-
Strategies and scheming: the war between PRRSV and host cells.Virol J. 2025 Jun 11;22(1):191. doi: 10.1186/s12985-025-02685-y. Virol J. 2025. PMID: 40500743 Free PMC article. Review.
-
Comparison of pathogenicity and host responses of emerging porcine reproductive and respiratory syndrome virus variants in piglets.J Virol. 2024 Nov 19;98(11):e0154223. doi: 10.1128/jvi.01542-23. Epub 2024 Oct 24. J Virol. 2024. PMID: 39445829 Free PMC article.
-
The role of immune checkpoint molecules in PRRSV-2-induced immune modulation: insights from comparative in vivo evaluation including NADC34-like PRRSV.J Virol. 2025 Jul 22;99(7):e0229824. doi: 10.1128/jvi.02298-24. Epub 2025 Jun 3. J Virol. 2025. PMID: 40459260 Free PMC article.
References
MeSH terms
Substances
Grants and funding
- ZR2021ZD08, R2020KC005, ZR2021MC139, ZR2023MC076/| Natural Science Foundation of Shandong Province ()
- 32373094, 32102710/MOST | National Natural Science Foundation of China (NSFC)
- CXGC2023A21, CXGC2023G03/the Agricultural Scienfitic and Technological Innovation Project of Shandong Academy of Agricultural Sciences
- 2018-67015-28287, 2023-67015-39710/U.S. Department of Agriculture (USDA)
LinkOut - more resources
Full Text Sources

