Multi-proteomic analyses of 5xFAD mice reveal new molecular signatures of early-stage Alzheimer's disease
- PMID: 38436501
- PMCID: PMC11166370
- DOI: 10.1111/acel.14137
Multi-proteomic analyses of 5xFAD mice reveal new molecular signatures of early-stage Alzheimer's disease
Abstract
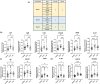
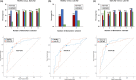
An early diagnosis of Alzheimer's disease is crucial as treatment efficacy is limited to the early stages. However, the current diagnostic methods are limited to mid or later stages of disease development owing to the limitations of clinical examinations and amyloid plaque imaging. Therefore, this study aimed to identify molecular signatures including blood plasma extracellular vesicle biomarker proteins associated with Alzheimer's disease to aid early-stage diagnosis. The hippocampus, cortex, and blood plasma extracellular vesicles of 3- and 6-month-old 5xFAD mice were analyzed using quantitative proteomics. Subsequent bioinformatics and biochemical analyses were performed to compare the molecular signatures between wild type and 5xFAD mice across different brain regions and age groups to elucidate disease pathology. There was a unique signature of significantly altered proteins in the hippocampal and cortical proteomes of 3- and 6-month-old mice. The plasma extracellular vesicle proteomes exhibited distinct informatic features compared with the other proteomes. Furthermore, the regulation of several canonical pathways (including phosphatidylinositol 3-kinase/protein kinase B signaling) differed between the hippocampus and cortex. Twelve potential biomarkers for the detection of early-stage Alzheimer's disease were identified and validated using plasma extracellular vesicles from stage-divided patients. Finally, integrin α-IIb, creatine kinase M-type, filamin C, glutamine γ-glutamyltransferase 2, and lysosomal α-mannosidase were selected as distinguishing biomarkers for healthy individuals and early-stage Alzheimer's disease patients using machine learning modeling with approximately 79% accuracy. Our study identified novel early-stage molecular signatures associated with the progression of Alzheimer's disease, thereby providing novel insights into its pathogenesis.
Keywords: Alzheimer's disease; biomarker; early‐stage Alzheimer's disease; extracellular vesicle; machine learning; proteomics.
© 2024 The Authors. Aging Cell published by Anatomical Society and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
-
- Adeoye, O. M. , Ferrell, R. E. , Kirshner, M. A. , Mulsant, B. H. , Seligman, K. , Begley, A. E. , Reynolds, C. F., 3rd , & Pollock, B. G. (2003). Alpha1‐acid glycoprotein in late‐life depression: Relationship to medical burden and genetics. Journal of Geriatric Psychiatry and Neurology, 16(4), 235–239. 10.1177/0891988703258321 - DOI - PubMed
-
- Aisen, P. S. , Cummings, J. , Jack, C. R., Jr. , Morris, J. C. , Sperling, R. , Frölich, L. , Jones, R. W. , Dowsett, S. A. , Matthews, B. R. , Raskin, J. , Scheltens, P. , & Dubois, B. (2017). On the path to 2025: Understanding the Alzheimer's disease continuum. Alzheimer's Research & Therapy, 9(1), 60. 10.1186/s13195-017-0283-5 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

