Cationic cholesterol-dependent LNP delivery to lung stem cells, the liver, and heart
- PMID: 38437539
- PMCID: PMC10945827
- DOI: 10.1073/pnas.2307801120
Cationic cholesterol-dependent LNP delivery to lung stem cells, the liver, and heart
Abstract
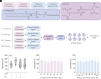
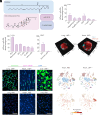
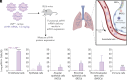
Adding a cationic helper lipid to a lipid nanoparticle (LNP) can increase lung delivery and decrease liver delivery. However, it remains unclear whether charge-dependent tropism is universal or, alternatively, whether it depends on the component that is charged. Here, we report evidence that cationic cholesterol-dependent tropism can differ from cationic helper lipid-dependent tropism. By testing how 196 LNPs delivered mRNA to 22 cell types, we found that charged cholesterols led to a different lung:liver delivery ratio than charged helper lipids. We also found that combining cationic cholesterol with a cationic helper lipid led to mRNA delivery in the heart as well as several lung cell types, including stem cell-like populations. These data highlight the utility of exploring charge-dependent LNP tropism.
Keywords: LNP; barcoding; mRNA; nanoparticle; scRNA-seq.
Conflict of interest statement
Competing interests statement:J.E.D. is an advisor to GV, Nava Therapeutics, and Edge Animal Health. All other authors declare no competing interests.
Figures


References
-
- Adams D., et al. , Patisiran, an RNAi therapeutic, for hereditary transthyretin amyloidosis. N. Engl. J. Med. 379, 11–21 (2018). - PubMed
-
- Gillmore J. D., et al. , CRISPR-Cas9 in vivo gene editing for transthyretin amyloidosis. N. Engl. J. Med. 385, 493–502 (2021). - PubMed
-
- Anonymous, Intellia therapeutics presents new interim data from first-in-human study of NTLA-2002 for the treatment of hereditary angioedema (HAE) at the American College of Allergy, Asthma & Immunology 2022 Annual Scientific Meeting. https://ir.intelliatx.com/news-releases/news-release-details/intellia-th... (2022). Accessed 8 June 2023.
-
- Loughrey D., Dahlman J. E., Non-liver mRNA delivery. Accounts Chem. Res. 55, 13–23 (2022). - PubMed
-
- Saunders N. R. M., et al. , A nanoprimer to improve the systemic delivery of siRNA and mRNA. Nano Lett. 20, 4264–4269 (2020). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

