Discovery of Red-Shifting Mutations in Firefly Luciferase Using High-Throughput Biochemistry
- PMID: 38437583
- PMCID: PMC10956436
- DOI: 10.1021/acs.biochem.3c00708
Discovery of Red-Shifting Mutations in Firefly Luciferase Using High-Throughput Biochemistry
Abstract
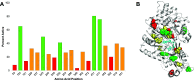
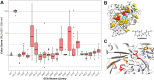
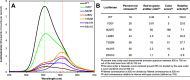
Photinus pyralis luciferase (FLuc) has proven a valuable tool for bioluminescence imaging, but much of the light emitted from the native enzyme is absorbed by endogenous biomolecules. Thus, luciferases displaying red-shifted emission enable higher resolution during deep-tissue imaging. A robust model of how protein structure determines emission color would greatly aid the engineering of red-shifted mutants, but no consensus has been reached to date. In this work, we applied deep mutational scanning to systematically assess 20 functionally important amino acid positions on FLuc for red-shifting mutations, predicting that an unbiased approach would enable novel contributions to this debate. We report dozens of red-shifting mutations as a result, a large majority of which have not been previously identified. Further characterization revealed that mutations N229T and T352M, in particular, bring about unimodal emission with the majority of photons being >600 nm. The red-shifting mutations identified by this high-throughput approach provide strong biochemical evidence for the multiple-emitter mechanism of color determination and point to the importance of a water network in the enzyme binding pocket for altering the emitter ratio. This work provides a broadly applicable mutational data set tying FLuc structure to emission color that contributes to our mechanistic understanding of emission color determination and should facilitate further engineering of improved probes for deep-tissue imaging.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

