A Pre-Leukemic DNA Methylation Signature in Healthy Individuals at Higher Risk for Developing Myeloid Malignancy
- PMID: 38437679
- PMCID: PMC11096012
- DOI: 10.1158/1078-0432.CCR-22-3804
A Pre-Leukemic DNA Methylation Signature in Healthy Individuals at Higher Risk for Developing Myeloid Malignancy
Abstract
Purpose: DNA methylation alterations are widespread in acute myelogenous leukemia (AML) and myelodysplastic syndrome (MDS), some of which appear to have evolved independently of somatic mutations in epigenetic regulators. Although the presence of somatic mutations in peripheral blood can predict the risk of development of AML and MDS, its accuracy remains unsatisfactory.
Experimental design: We performed global DNA methylation profiling in a case control study nested within the Singapore Chinese Health Study to evaluate whether DNA methylation alterations were associated with AML/MDS development. Targeted deep sequencing and methylated DNA immunoprecipitation sequencing (MeDIP-seq) were performed on peripheral blood collected a median of 9.9 years before diagnosis of AML or MDS, together with age-matched still-healthy individuals as controls.
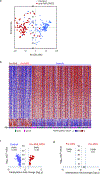
Results: Sixty-six individuals who developed AML or MDS displayed significant DNA methylation changes in the peripheral blood compared with 167 age- and gender-matched controls who did not develop AML/MDS during the follow-up period. Alterations in methylation in the differentially methylation regions were associated with increased odds of developing AML/MDS.
Conclusions: The epigenetic changes may be acquired independently and before somatic mutations that are relevant for AML/MDS development. The association between methylation changes and the risk of pre-AML/MDS in these individuals was considerably stronger than somatic mutations, suggesting that methylation changes could be used as biomarkers for pre-AML/MDS screening.
©2024 American Association for Cancer Research.
Conflict of interest statement
Figures



Similar articles
-
Genetic variants involved in oxidative stress, base excision repair, DNA methylation, and folate metabolism pathways influence myeloid neoplasias susceptibility and prognosis.Mol Carcinog. 2017 Jan;56(1):130-148. doi: 10.1002/mc.22478. Epub 2016 Mar 7. Mol Carcinog. 2017. PMID: 26950655
-
Epigenetic modifications of splicing factor genes in myelodysplastic syndromes and acute myeloid leukemia.Cancer Sci. 2014 Nov;105(11):1457-63. doi: 10.1111/cas.12532. Epub 2014 Oct 18. Cancer Sci. 2014. PMID: 25220401 Free PMC article.
-
Promoter methylation of DAPK1, E-cadherin and thrombospondin-1 in de novo and therapy-related myeloid neoplasms.Blood Cells Mol Dis. 2010 Oct 15;45(3):181-5. doi: 10.1016/j.bcmd.2010.05.008. Epub 2010 Jul 24. Blood Cells Mol Dis. 2010. PMID: 20655775
-
The changing mutational landscape of acute myeloid leukemia and myelodysplastic syndrome.Mol Cancer Res. 2013 Aug;11(8):815-27. doi: 10.1158/1541-7786.MCR-12-0695. Epub 2013 May 3. Mol Cancer Res. 2013. PMID: 23645565 Free PMC article. Review.
-
Universal genetic testing for inherited susceptibility in children and adults with myelodysplastic syndrome and acute myeloid leukemia: are we there yet?Leukemia. 2018 Jul;32(7):1482-1492. doi: 10.1038/s41375-018-0051-y. Epub 2018 Feb 27. Leukemia. 2018. PMID: 29483711 Review.
Cited by
-
Epigenomic insights and computational advances in hematologic malignancies.Mol Cytogenet. 2025 Apr 12;18(1):9. doi: 10.1186/s13039-025-00712-9. Mol Cytogenet. 2025. PMID: 40221777 Free PMC article. Review.
References
-
- Shlush LI. Age-related clonal hematopoiesis. Blood 2018;131(5):496–504. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

