Zα Domain of ADAR1 Binds to an A-Form-like Nucleic Acid Duplex with Low Micromolar Affinity
- PMID: 38437710
- PMCID: PMC11168418
- DOI: 10.1021/acs.biochem.3c00636
Zα Domain of ADAR1 Binds to an A-Form-like Nucleic Acid Duplex with Low Micromolar Affinity
Abstract
The left-handed Z-conformation of nucleic acids can be adopted by both DNA and RNA when bound by Zα domains found within a variety of viral and innate immune response proteins. While Z-form adoption is preferred by certain sequences, such as the commonly studied (CpG)n repeats, Zα has been reported to bind to a wide range of sequence contexts. Studying how Zα interacts with B-/A-form helices prior to their conversion to the Z-conformation is challenging as binding coincides with Z-form adoption. Here, we studied the binding of Zα fromHomo sapiens ADAR1 to a locked "A-type" version of the (CpG)3 construct (LNA (CpG)3) where the sugar pucker is locked into the C3'-endo/C2'-exo conformation, which prevents the duplex from adopting the alternating C2'/C3'-endo sugar puckers found in the Z-conformation. Using NMR and other biophysical techniques, we find that ZαADAR1 binds to the LNA (CpG)3 using a similar interface as for Z-form binding, with a dissociation constant (KD) of ∼4 μM. In contrast to Z-DNA/Z-RNA, where two ZαADAR1 bind to every 6 bp stretch, our data suggests that ZαADAR1 binds to multiple LNA molecules, indicating a completely different binding mode. Because ZαADAR1 binds relatively tightly to a non-Z-form model, its binding to B/A-form helices may need to be considered when experiments are carried out which attempt to identify the Z-form targets of Zα domains. The use of LNA constructs may be beneficial in experiments where negative controls for Z-form adoption are needed.
Figures


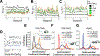
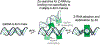
Similar articles
-
NMR investigation on the DNA binding and B-Z transition pathway of the Zα domain of human ADAR1.Biophys Chem. 2013 Feb;172:18-25. doi: 10.1016/j.bpc.2012.12.002. Epub 2012 Dec 21. Biophys Chem. 2013. PMID: 23334429
-
Thermodynamic analysis of Zα domain-nucleic acid interactions.Biochem J. 2022 Aug 31;479(16):1727-1741. doi: 10.1042/BCJ20220200. Biochem J. 2022. PMID: 35969150
-
Sequence discrimination of the Zα domain of human ADAR1 during B-Z transition of DNA duplexes.FEBS Lett. 2010 Oct 22;584(20):4344-50. doi: 10.1016/j.febslet.2010.09.036. Epub 2010 Sep 26. FEBS Lett. 2010. PMID: 20875819
-
Zalpha-domains: at the intersection between RNA editing and innate immunity.Semin Cell Dev Biol. 2012 May;23(3):275-80. doi: 10.1016/j.semcdb.2011.11.001. Epub 2011 Nov 7. Semin Cell Dev Biol. 2012. PMID: 22085847 Review.
-
Zα and Zβ Localize ADAR1 to Flipons That Modulate Innate Immunity, Alternative Splicing, and Nonsynonymous RNA Editing.Int J Mol Sci. 2025 Mar 7;26(6):2422. doi: 10.3390/ijms26062422. Int J Mol Sci. 2025. PMID: 40141064 Free PMC article. Review.
References
-
- Wolfram S Principles of Nucleic Acid Structure, 1st ed.; Springer New York: New York, 1984.
-
- Plavee J; Tong W; Chattopadhyaya J How Do the Gauche and Anomeric Effects Drive the Pseudorotational Equilibrium of the Pentofuranose Moiety of Nucleosides. J. Am. Chem. Soc. 1993. 10.1021/ja00074a046. - DOI
-
- Plavec J; Thibaudeau C; Chattopadhyaya J How Does the 2’-Hydroxy Group Drive the Pseudorotational Equilibrium in Nucleoside and Nucleotide by the Tuning of the 3’-Gauche Effect? J. Am. Chem. Soc. 1994. 10.1021/ja00094a009. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

