Altered myocardial lipid regulation in junctophilin-2-associated familial cardiomyopathies
- PMID: 38438248
- PMCID: PMC10912815
- DOI: 10.26508/lsa.202302330
Altered myocardial lipid regulation in junctophilin-2-associated familial cardiomyopathies
Abstract
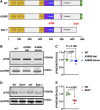
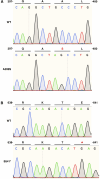
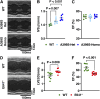
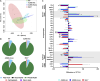
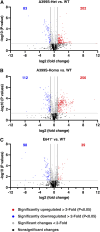
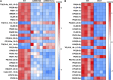
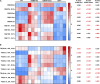
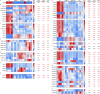
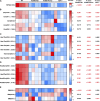
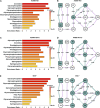
Myocardial lipid metabolism is critical to normal heart function, whereas altered lipid regulation has been linked to cardiac diseases including cardiomyopathies. Genetic variants in the JPH2 gene can cause hypertrophic cardiomyopathy (HCM) and, in some cases, dilated cardiomyopathy (DCM). In this study, we tested the hypothesis that JPH2 variants identified in patients with HCM and DCM, respectively, cause distinct alterations in myocardial lipid profiles. Echocardiography revealed clinically significant cardiac dysfunction in both knock-in mouse models of cardiomyopathy. Unbiased myocardial lipidomic analysis demonstrated significantly reduced levels of total unsaturated fatty acids, ceramides, and various phospholipids in both mice with HCM and DCM, suggesting a common metabolic alteration in both models. On the contrary, significantly increased di- and triglycerides, and decreased co-enzyme were only found in mice with HCM. Moreover, mice with DCM uniquely exhibited elevated levels of cholesterol ester. Further in-depth analysis revealed significantly altered metabolites from all the lipid classes with either similar or opposing trends in JPH2 mutant mice with HCM or DCM. Together, these studies revealed, for the first time, unique alterations in the cardiac lipid composition-including distinct increases in neutral lipids and decreases in polar membrane lipids-in mice with HCM and DCM were caused by distinct JPH2 variants. These studies may aid the development of novel biomarkers or therapeutics for these inherited disorders.
© 2024 Lahiri et al.
Conflict of interest statement
XHT Wehrens serves as a consultant for Pfizer and Rocket Pharmaceuticals, in addition to being a founding partner and board member at Elex Biotech Inc. The remaining authors have no relevant disclosures to report.
Figures



Similar articles
-
Mutations in JPH2-encoded junctophilin-2 associated with hypertrophic cardiomyopathy in humans.J Mol Cell Cardiol. 2007 Jun;42(6):1026-35. doi: 10.1016/j.yjmcc.2007.04.006. Epub 2007 Apr 18. J Mol Cell Cardiol. 2007. PMID: 17509612 Free PMC article.
-
Analysis of enriched rare variants in JPH2-encoded junctophilin-2 among Greater Middle Eastern individuals reveals a novel homozygous variant associated with neonatal dilated cardiomyopathy.Sci Rep. 2019 Jun 21;9(1):9038. doi: 10.1038/s41598-019-44987-6. Sci Rep. 2019. PMID: 31227780 Free PMC article.
-
Genetic evaluation of cardiomyopathies in Qatar identifies enrichment of pathogenic sarcomere gene variants and possible founder disease mutations in the Arabs.Mol Genet Genomic Med. 2021 Jul;9(7):e1709. doi: 10.1002/mgg3.1709. Epub 2021 Jun 17. Mol Genet Genomic Med. 2021. PMID: 34137518 Free PMC article.
-
Inflammatory dilated cardiomyopathy (DCMI).Herz. 2005 Sep;30(6):535-44. doi: 10.1007/s00059-005-2730-5. Herz. 2005. PMID: 16170686 Review.
-
Dilated cardiomyopathy: a disease of the myocardium.Biol Pharm Bull. 2013;36(1):18-22. doi: 10.1248/bpb.b212023. Biol Pharm Bull. 2013. PMID: 23302633 Review.
Cited by
-
Evaluating the functional and genomic analysis of pathogenic junctophilin-2 variants and their association with the pathogenesis of cardiomyopathy to understand their molecular impact on cardiac calcium homeostasis and disease phenotypes.Ann Pediatr Cardiol. 2024 Nov-Dec;17(6):401-407. doi: 10.4103/apc.apc_173_24. Epub 2025 Apr 24. Ann Pediatr Cardiol. 2024. PMID: 40352421 Free PMC article. Review.
-
Unveiling the Spectrum of Minor Genes in Cardiomyopathies: A Narrative Review.Int J Mol Sci. 2024 Sep 10;25(18):9787. doi: 10.3390/ijms25189787. Int J Mol Sci. 2024. PMID: 39337275 Free PMC article. Review.
References
-
- Arimoto T, Takeishi Y, Takahashi H, Shishido T, Niizeki T, Koyama Y, Shiga R, Nozaki N, Nakajima O, Nishimaru K, et al. (2006) Cardiac-specific overexpression of diacylglycerol kinase zeta prevents Gq protein-coupled receptor agonist-induced cardiac hypertrophy in transgenic mice. Circulation 113: 60–66. 10.1161/CIRCULATIONAHA.105.560771 - DOI - PubMed
-
- Bennett HJ, Davenport JB, Collins RF, Trafford AW, Pinali C, Kitmitto A (2013) Human junctophilin-2 undergoes a structural rearrangement upon binding PtdIns(3,4,5)P3 and the S101R mutation identified in hypertrophic cardiomyopathy obviates this response. Biochem J 456: 205–217. 10.1042/BJ20130591 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
