RANBP17 Overexpression Restores Nucleocytoplasmic Transport and Ameliorates Neurodevelopment in Induced DYT1 Dystonia Motor Neurons
- PMID: 38438257
- PMCID: PMC11007476
- DOI: 10.1523/JNEUROSCI.1728-23.2024
RANBP17 Overexpression Restores Nucleocytoplasmic Transport and Ameliorates Neurodevelopment in Induced DYT1 Dystonia Motor Neurons
Abstract
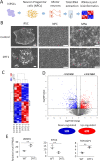
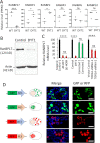
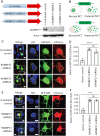
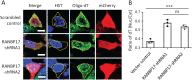
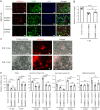
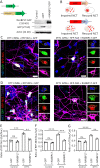
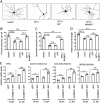
DYT1 dystonia is a debilitating neurological movement disorder, and it represents the most frequent and severe form of hereditary primary dystonia. There is currently no cure for this disease due to its unclear pathogenesis. In our previous study utilizing patient-specific motor neurons (MNs), we identified distinct cellular deficits associated with the disease, including a deformed nucleus, disrupted neurodevelopment, and compromised nucleocytoplasmic transport (NCT) functions. However, the precise molecular mechanisms underlying these cellular impairments have remained elusive. In this study, we revealed the genome-wide changes in gene expression in DYT1 MNs through transcriptomic analysis. We found that those dysregulated genes are intricately involved in neurodevelopment and various biological processes. Interestingly, we identified that the expression level of RANBP17, a RAN-binding protein crucial for NCT regulation, exhibited a significant reduction in DYT1 MNs. By manipulating RANBP17 expression, we further demonstrated that RANBP17 plays an important role in facilitating the nuclear transport of both protein and transcript cargos in induced human neurons. Excitingly, the overexpression of RANBP17 emerged as a substantial mitigating factor, effectively restoring impaired NCT activity and rescuing neurodevelopmental deficits observed in DYT1 MNs. These findings shed light on the intricate molecular underpinnings of impaired NCT in DYT1 neurons and provide novel insights into the pathophysiology of DYT1 dystonia, potentially leading to the development of innovative treatment strategies.
Keywords: RANBP17; Torsin ATPase; dystonia; human-induced pluripotent stem cells (hiPSCs); motor neurons; neurodevelopment; nucleocytoplasmic transport.
Copyright © 2024 the authors.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

Similar articles
-
Disease Modeling with Human Neurons Reveals LMNB1 Dysregulation Underlying DYT1 Dystonia.J Neurosci. 2021 Mar 3;41(9):2024-2038. doi: 10.1523/JNEUROSCI.2507-20.2020. Epub 2021 Jan 19. J Neurosci. 2021. PMID: 33468570 Free PMC article.
-
Cell-specific effects of Dyt1 knock-out on sensory processing, network-level connectivity, and motor deficits.Exp Neurol. 2021 Sep;343:113783. doi: 10.1016/j.expneurol.2021.113783. Epub 2021 Jun 10. Exp Neurol. 2021. PMID: 34119482 Free PMC article.
-
Hypertrophy of nigral neurons in Torsin1A deletion (DYT1) carriers manifesting dystonia.Parkinsonism Relat Disord. 2019 Jan;58:63-69. doi: 10.1016/j.parkreldis.2018.08.020. Epub 2018 Aug 31. Parkinsonism Relat Disord. 2019. PMID: 30193818
-
TorsinA and DYT1 dystonia: a synaptopathy?Biochem Soc Trans. 2010 Apr;38(2):452-6. doi: 10.1042/BST0380452. Biochem Soc Trans. 2010. PMID: 20298201 Review.
-
The cognitive features of idiopathic and DYT1 dystonia.Mov Disord. 2017 Oct;32(10):1348-1355. doi: 10.1002/mds.27048. Epub 2017 Jun 19. Mov Disord. 2017. PMID: 28627117 Review.
Cited by
-
Neurobiology of Dystonia: Review of Genetics, Animal Models, and Neuroimaging.Brain Sci. 2025 Jul 19;15(7):767. doi: 10.3390/brainsci15070767. Brain Sci. 2025. PMID: 40722357 Free PMC article. Review.
-
Assembling a Coculture System to Prepare Highly Pure Induced Pluripotent Stem Cell-Derived Neurons at Late Maturation Stages.eNeuro. 2024 Jul 30;11(7):ENEURO.0165-24.2024. doi: 10.1523/ENEURO.0165-24.2024. Print 2024 Jul. eNeuro. 2024. PMID: 39009447 Free PMC article.
-
Deciphering the Pathophysiological Mechanisms Underpinning Myoclonus Dystonia Using Pluripotent Stem Cell-Derived Cellular Models.Cells. 2024 Sep 10;13(18):1520. doi: 10.3390/cells13181520. Cells. 2024. PMID: 39329704 Free PMC article. Review.
References
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
